For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSTQIRKRARPRSLVVAGLLPLAGLLGVITTPAYATSADDAFLAALKAKGINYESPDAAVNSGHTVCHE LDMGQTPEQVANNVLSSSTLDSYHAGYFVGVSIKAYCPKYAAAAAGS
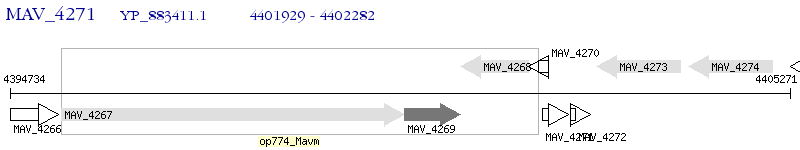
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4271 | - | - | 100% (117) | hypothetical protein MAV_4271 |
| M. avium 104 | MAV_3549 | - | 3e-14 | 37.27% (110) | hypothetical protein MAV_3549 |
| M. avium 104 | MAV_3558 | - | 1e-12 | 38.00% (100) | hypothetical protein MAV_3558 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1302c | - | 1e-13 | 37.50% (88) | hypothetical protein Mb1302c |
| M. gilvum PYR-GCK | Mflv_2012 | - | 2e-11 | 32.95% (88) | hypothetical protein Mflv_2012 |
| M. tuberculosis H37Rv | Rv1271c | - | 1e-13 | 37.50% (88) | hypothetical protein Rv1271c |
| M. leprae Br4923 | MLBr_00676 | - | 7e-08 | 30.28% (109) | hypothetical protein MLBr_00676 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1233 | - | 2e-24 | 48.60% (107) | hypothetical protein MMAR_1233 |
| M. smegmatis MC2 155 | MSMEG_4689 | - | 8e-08 | 34.86% (109) | hypothetical protein MSMEG_4689 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2657 | - | 1e-24 | 48.60% (107) | hypothetical protein MUL_2657 |
| M. vanbaalenii PYR-1 | Mvan_4716 | - | 3e-13 | 31.91% (94) | hypothetical protein Mvan_4716 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1233|M.marinum_M --------------------------------------------------
MUL_2657|M.ulcerans_Agy99 MQRSVPAKPQRGRIRKFSHSLRPVHAPERRPTSPRPDGQLQPVAGMAAEG
MAV_4271|M.avium_104 ---------------------------------------------MTSTQ
Mb1302c|M.bovis_AF2122/97 --------------------------------------------------
Rv1271c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_2012|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4716|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00676|M.leprae_Br4923 -----------------------------MFLRLMLSALKALQRLGAVMN
MSMEG_4689|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1233|M.marinum_M -----------MRSSRTRALVLV--VALVGLTELATAT-AHADAVDDAFL
MUL_2657|M.ulcerans_Agy99 RRALLGGEVPAMRSSRTRALVLV--VALVGLTELATAT-AHADAVDDAFL
MAV_4271|M.avium_104 IR----------KRARPRSLVVAGLLPLAGLLGVITTP-AYATSADDAFL
Mb1302c|M.bovis_AF2122/97 -------MLSPLSPRIIAAFTTAVGAAAIGLAVATAGT-AGANTKDEAFI
Rv1271c|M.tuberculosis_H37Rv -------MLSPLSPRIIAAFTTAVGAAAIGLAVATAGT-AGANTKDEAFI
Mflv_2012|M.gilvum_PYR-GCK ---------------MRGTFFITT-LAAAGLATALAAP-AFADETDDIFI
Mvan_4716|M.vanbaalenii_PYR-1 ---------------MRGKLIVSAAVAAAGLATALAAP-AFADETDDIFI
MLBr_00676|M.leprae_Br4923 SLARIDHWIWLFRCQPLTIRLLVATAALFTAATAFEVP-AEADAIDDTFI
MSMEG_4689|M.smegmatis_MC2_155 -------MSARLPAVLVTAAALAAGVGATPAHADPAAPGAPGNPVDRSFL
. * . * *:
MMAR_1233|M.marinum_M NAVRSHGINFASPQAAIMAGHQVCNALD-SGRQKSDVAGDVAAS-SSLDG
MUL_2657|M.ulcerans_Agy99 NAVRSHGINFASPQAAIMAGHQVCNALD-SGRQKSDVAGDVAAS-SSLDG
MAV_4271|M.avium_104 AALKAKGINYESPDAAVNSGHTVCHELD-MGQTPEQVANNVLSS-STLDS
Mb1302c|M.bovis_AF2122/97 AQMESIGVTFSSPQVATQQAQLVCKKLA-SGETGTEIAEEVLSQ-TNLTT
Rv1271c|M.tuberculosis_H37Rv AQMESIGVTFSSPQVATQQAQLVCKKLA-SGETGTEIAEEVLSQ-TNLTT
Mflv_2012|M.gilvum_PYR-GCK AALEQEGIPFSTAGNAIELAGAVCEYAA-AGQDKTQIALEIMEP-AGWSP
Mvan_4716|M.vanbaalenii_PYR-1 SALQNQGIPFSTAENAIELASAVCDYAA-AGQDPTQIALEIMEP-AGWTA
MLBr_00676|M.leprae_Br4923 KALNHAGVNFGEPRSAMTMGHYVCPILAKSGGNFAAAVQRIRGN-SDMSP
MSMEG_4689|M.smegmatis_MC2_155 TALNNAGVGYADPDAAVRLGQDVCPMLVEPGKNFASVVTKLRGDRSVVSP
:. *: : . * . ** * . : :
MMAR_1233|M.marinum_M YRAGYFVGVSIAAYCPRHHSQA-------------------
MUL_2657|M.ulcerans_Agy99 YRAGYFVGVSIAAYCPRHHSQA-------------------
MAV_4271|M.avium_104 YHAGYFVGVSIKAYCPKYAAAAAGS----------------
Mb1302c|M.bovis_AF2122/97 KQAAYFVVDATKAYCPQYASQLT------------------
Rv1271c|M.tuberculosis_H37Rv KQAAYFVVDATKAYCPQYASQLT------------------
Mflv_2012|M.gilvum_PYR-GCK EQSGFFVGAATQSYCP-------------------------
Mvan_4716|M.vanbaalenii_PYR-1 EQSGFFVGAATQSYCPS------------------------
MLBr_00676|M.leprae_Br4923 QMAETFAKIAISIYCPTMMANVASGNLPSLPPGPGIPGI--
MSMEG_4689|M.smegmatis_MC2_155 DVAAFFAGIAISMYCPQMISGIGDGTLLRMLAVPGLGALGR
: *. : ***