For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGLHNHRLVLLRHGETEWSKTGRHTGRTEVELTEAGRTQAERAGRALSELKLVEPLVISSPRQRSLATAE LAGLNVDEVSEQLAEWDYGSYEGLTTEQIRQSVPDWLVWTHGCPGGESVAQVSGRADAAVTAALRQMESR DVVFVSHGHFCRAVMTRWVELPLAEGSRFAMITASIAVCGFEHGVRQLRALGLT
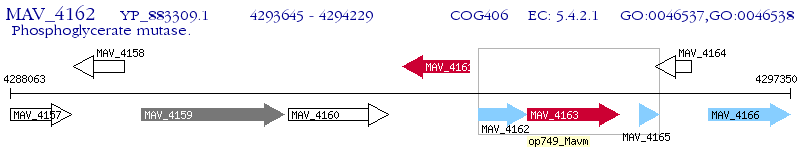
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4162 | - | - | 100% (194) | acid phosphatase |
| M. avium 104 | MAV_2211 | - | 9e-16 | 38.67% (150) | bifunctional RNase H/acid phosphatase |
| M. avium 104 | MAV_1752 | - | 2e-13 | 33.71% (175) | phosphoglycerate mutase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3240 | gpm2 | 3e-83 | 75.26% (194) | acid phosphatase |
| M. gilvum PYR-GCK | Mflv_4679 | - | 3e-74 | 67.01% (194) | acid phosphatase |
| M. tuberculosis H37Rv | Rv3214 | gpm2 | 3e-83 | 75.26% (194) | acid phosphatase |
| M. leprae Br4923 | MLBr_00808 | entC | 6e-26 | 59.78% (92) | putative isochorismate synthase |
| M. abscessus ATCC 19977 | MAB_3534 | - | 1e-72 | 66.67% (189) | acid phosphatase |
| M. marinum M | MMAR_1343 | gpm2 | 6e-84 | 75.65% (193) | phosphoglycerate mutase Gpm2 |
| M. smegmatis MC2 155 | MSMEG_1926 | - | 4e-75 | 67.53% (194) | acid phosphatase |
| M. thermoresistible (build 8) | TH_0648 | entD | 3e-73 | 69.35% (186) | POSSIBLE PHOSPHOGLYCERATE MUTASE GPM2 (PHOSPHOGLYCEROMUTASE) |
| M. ulcerans Agy99 | MUL_2536 | gpm2 | 1e-83 | 75.65% (193) | acid phosphatase |
| M. vanbaalenii PYR-1 | Mvan_1788 | - | 7e-74 | 66.49% (194) | acid phosphatase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3240|M.bovis_AF2122/97 -------------------------MGVRNHRLLLLRHGETAWSTLGRHT
Rv3214|M.tuberculosis_H37Rv -------------------------MGVRNHRLLLLRHGETAWSTLGRHT
MMAR_1343|M.marinum_M -------------MGPACRPAQSFGMGVHNHRLLLLRHGDTEWSKSGKHT
MUL_2536|M.ulcerans_Agy99 MSSWHVLVSLAGAMGPACRPAQSFGMGVHNHRLLLLRHGDTEWSKSGKHT
MAV_4162|M.avium_104 -------------------------MGLHNHRLVLLRHGETEWSKTGRHT
MSMEG_1926|M.smegmatis_MC2_155 -------------------------MSVQQHRLLLLRHGETEWSASGRHT
TH_0648|M.thermoresistible__bu ---------------------------------VLLRHGETEWSRSGRHT
Mflv_4679|M.gilvum_PYR-GCK -MALPCACSPARSCDKPRRTAESDSVGLLQHRLILLRHGETEWSKSGKHT
Mvan_1788|M.vanbaalenii_PYR-1 -------------------------MGLLQHRLILLRHGETEWSKSGKHT
MAB_3534|M.abscessus_ATCC_1997 -------------------------MSALLNRMLLLRHGETEWSKIGRHT
MLBr_00808|M.leprae_Br4923 ---------------------------------MAIRRGRNRASTFG---
: :*:* . * *
Mb3240|M.bovis_AF2122/97 GGTEVELTDTGRTQAELAGQLLGELELDDPIVICSPRRRTLDTAKLAGLT
Rv3214|M.tuberculosis_H37Rv GGTEVELTDTGRTQAELAGQLLGELELDDPIVICSPRRRTLDTAKLAGLT
MMAR_1343|M.marinum_M GRTDIELTETGRAQAELAGRALNELELDNPLVISSPRKRTLATAELAGLT
MUL_2536|M.ulcerans_Agy99 GRTDIELTETGRAQAELAGRALNELELDNPLVISSPRKRTLATAELAGLT
MAV_4162|M.avium_104 GRTEVELTEAGRTQAERAGRALSELKLVEPLVISSPRQRSLATAELAGLN
MSMEG_1926|M.smegmatis_MC2_155 GRTDLDLTEAGREQAKLAAEPLAELQLRDPLVFSSPRRRALVTAELAGLT
TH_0648|M.thermoresistible__bu GRTDLELTETGRNRAKLAAEPLAALGLDNPLVVSSPMRRARDTAELAGLR
Mflv_4679|M.gilvum_PYR-GCK SSTELELTEHGRAQARAAADTLKQMELDDPYVVSSPRVRATTTADLAGLT
Mvan_1788|M.vanbaalenii_PYR-1 SRTELELTEHGREQALAAADTLKQMQLQDPFVVTSPRIRARTTAELAGLV
MAB_3534|M.abscessus_ATCC_1997 GRTDLDLTEIGRAQAVAAREVLKDLDLNDPLVLSSPRRRALQTAELAGLT
MLBr_00808|M.leprae_Br4923 -CTELELTDTGRAQAKLAERALAEQKLVDPMMASNPRQCDLVTAKLAGLT
*:::**: ** :* * * * :* : .* **.****
Mb3240|M.bovis_AF2122/97 VNEVTGLLAEWDYGSYEGLTTPQIRESEPDWLVWTHGCPAGESVAQVNDR
Rv3214|M.tuberculosis_H37Rv VNEVTGLLAEWDYGSYEGLTTPQIRESEPDWLVWTHGCPAGESVAQVNDR
MMAR_1343|M.marinum_M VDTVAELVAEWDYGSYEGLTTPQIREFEPNWLVWTHGCPGGESVQQVTER
MUL_2536|M.ulcerans_Agy99 VDTVSELVAEWDYGSYEGLITPQIREFEPNWLVWTHGCPGGESVQQVTER
MAV_4162|M.avium_104 VDEVSEQLAEWDYGSYEGLTTEQIRQSVPDWLVWTHGCPGGESVAQVSGR
MSMEG_1926|M.smegmatis_MC2_155 VAEELPLLAEWDYGNYEGLTTAQIRETEPDWLVWKHGCPGGESVEEVGER
TH_0648|M.thermoresistible__bu VAEVSELLHEWDYGEYEGLTTAEIRRSVPDWLVWTHGCPGGESVADVGAR
Mflv_4679|M.gilvum_PYR-GCK VDEVSPLVSEWDYGDYEGTTTADIRKAVPNWLVWTHGCPGGETSAQVCAR
Mvan_1788|M.vanbaalenii_PYR-1 IDEVDDLIAEWDYGDYEGTTTADIRKAVPNWLVWTHGCPGGETSAQVCER
MAB_3534|M.abscessus_ATCC_1997 VDTVDEQLAEWDYGDYEGLTTPQIRENDPGWLVWTHGCPGGESVAQVQAR
MLBr_00808|M.leprae_Br4923 VDKVSPLLAECTYGSYEDLTTAKTREAEPAWLAWTHRYPRGEKRLASQRT
: : * **.**. * . *. * **.*.* * **.
Mb3240|M.bovis_AF2122/97 ADSAVALALEHMSSRD--------VLFVSHGHFSRAVITR----------
Rv3214|M.tuberculosis_H37Rv ADSAVALALEHMSSRD--------VLFVSHGHFSRAVITR----------
MMAR_1343|M.marinum_M ADRAIALALEHMTSRD--------VVFASHSHFSRSVITR----------
MUL_2536|M.ulcerans_Agy99 ADRAIALALEHMTSRD--------VVFASHSHFSRSVTTR----------
MAV_4162|M.avium_104 ADAAVTAALRQMESRD--------VVFVSHGHFCRAVMTR----------
MSMEG_1926|M.smegmatis_MC2_155 ADRAVALALEYLEDRD--------VVFVGHGHFSRAVLAR----------
TH_0648|M.thermoresistible__bu ADRALAYATEQLAHRD--------VVFVGHGHFSRAMITR----------
Mflv_4679|M.gilvum_PYR-GCK ADQAIDVALGHMESRD--------VVFVGHGHFSRAVITR----------
Mvan_1788|M.vanbaalenii_PYR-1 ADRAIAVALQHMQSRD--------VVFVGHGNFSRAVMTR----------
MAB_3534|M.abscessus_ATCC_1997 ADLVITKIAAELTQRD--------VLLVGHGHFSRALMTR----------
MLBr_00808|M.leprae_Br4923 CRPGRRYGVRALGVTDHVVRQPWPPLLGGDHTLDRAAICRGPALRYAHQL
. : * :: .. : *: *
Mb3240|M.bovis_AF2122/97 ----------------WVQLPLAEG---------------------SRFA
Rv3214|M.tuberculosis_H37Rv ----------------WVQLPLAEG---------------------SRFA
MMAR_1343|M.marinum_M ----------------WVELALVEG---------------------SRFY
MUL_2536|M.ulcerans_Agy99 ----------------WVELALVEG---------------------SRFY
MAV_4162|M.avium_104 ----------------WVELPLAEG---------------------SRFA
MSMEG_1926|M.smegmatis_MC2_155 ----------------WVELPVTHG---------------------IRIS
TH_0648|M.thermoresistible__bu ----------------WAELPVSQG---------------------IRFA
Mflv_4679|M.gilvum_PYR-GCK ----------------WIEQPVYEG---------------------IRFA
Mvan_1788|M.vanbaalenii_PYR-1 ----------------WIEQPVYEG---------------------IRFA
MAB_3534|M.abscessus_ATCC_1997 ----------------WIDLPLEEG---------------------TRLG
MLBr_00808|M.leprae_Br4923 DWDVRVRVRATATTHAWVAWLATTGRTRVNINNDDDKRRPVAQERAEQMR
* * ::
Mb3240|M.bovis_AF2122/97 MPTASIGICG-----FEHGVRQ--------------------LAVLGLTG
Rv3214|M.tuberculosis_H37Rv MPTASIGICG-----FEHGVRQ--------------------LAVLGLTG
MMAR_1343|M.marinum_M MPTASIAICG-----FEHGVRQ--------------------LSALGLKG
MUL_2536|M.ulcerans_Agy99 MPTASIAICG-----FEHGVRQ--------------------LSALGLKG
MAV_4162|M.avium_104 MITASIAVCG-----FEHGVRQ--------------------LRALGLT-
MSMEG_1926|M.smegmatis_MC2_155 MVPASIGVCG-----FEHGVRQ--------------------ISALGLTG
TH_0648|M.thermoresistible__bu MTPAAIAVCG-----FEHGVRQ--------------------VGALGLTG
Mflv_4679|M.gilvum_PYR-GCK MPAASIAVCG-----FEHGVRQ--------------------LSALAQTG
Mvan_1788|M.vanbaalenii_PYR-1 MPAASIAVCG-----FEHGVRQ--------------------LSALAQTG
MAB_3534|M.abscessus_ATCC_1997 MATASIAIGG-----HEHGQRQ--------------------VAALGLTG
MLBr_00808|M.leprae_Br4923 PSEPPFALCGPRGTLVADGVHTRYDDVYLAQAALRSGVTPILLGALPFDA
..:.: * .* : : .*
Mb3240|M.bovis_AF2122/97 HPQPIAAG------------------------------------------
Rv3214|M.tuberculosis_H37Rv HPQPIAAG------------------------------------------
MMAR_1343|M.marinum_M FSDS----------------------------------------------
MUL_2536|M.ulcerans_Agy99 FSDS----------------------------------------------
MAV_4162|M.avium_104 --------------------------------------------------
MSMEG_1926|M.smegmatis_MC2_155 HPNPCLPQ------------------------------------------
TH_0648|M.thermoresistible__bu YQHPDR--------------------------------------------
Mflv_4679|M.gilvum_PYR-GCK LPNPAVST------------------------------------------
Mvan_1788|M.vanbaalenii_PYR-1 RPNPAVST------------------------------------------
MAB_3534|M.abscessus_ATCC_1997 YQHPVPSS------------------------------------------
MLBr_00808|M.leprae_Br4923 ESPAALMAPSAVLRTDTLPNWQTGPLPTLRITAAVPTRANYRVWISNARE
Mb3240|M.bovis_AF2122/97 --------------------------------------------------
Rv3214|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1343|M.marinum_M --------------------------------------------------
MUL_2536|M.ulcerans_Agy99 --------------------------------------------------
MAV_4162|M.avium_104 --------------------------------------------------
MSMEG_1926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0648|M.thermoresistible__bu --------------------------------------------------
Mflv_4679|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1788|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3534|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00808|M.leprae_Br4923 QLTAPGNSLHKVVLARALQLVADAPLDGRAILRRLVTADPTAYGYLVDLT
Mb3240|M.bovis_AF2122/97 --------------------------------------------------
Rv3214|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1343|M.marinum_M --------------------------------------------------
MUL_2536|M.ulcerans_Agy99 --------------------------------------------------
MAV_4162|M.avium_104 --------------------------------------------------
MSMEG_1926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0648|M.thermoresistible__bu --------------------------------------------------
Mflv_4679|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1788|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3534|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00808|M.leprae_Br4923 AAGGQYAGVALVGASPELLVSRFGDRVVCRPFAGSAPRATNPEVDSANSA
Mb3240|M.bovis_AF2122/97 --------------------------------------------------
Rv3214|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1343|M.marinum_M --------------------------------------------------
MUL_2536|M.ulcerans_Agy99 --------------------------------------------------
MAV_4162|M.avium_104 --------------------------------------------------
MSMEG_1926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0648|M.thermoresistible__bu --------------------------------------------------
Mflv_4679|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1788|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3534|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00808|M.leprae_Br4923 ALAGSIKNRHEHQLVIDAIRAALEPVCDNLTIAPEPELSRTAAVWHLCTP
Mb3240|M.bovis_AF2122/97 --------------------------------------------------
Rv3214|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1343|M.marinum_M --------------------------------------------------
MUL_2536|M.ulcerans_Agy99 --------------------------------------------------
MAV_4162|M.avium_104 --------------------------------------------------
MSMEG_1926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0648|M.thermoresistible__bu --------------------------------------------------
Mflv_4679|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1788|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3534|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00808|M.leprae_Br4923 ISGRLRDTSITAIDLALMLHPTPAVGGVPAEAASELIAALEGDRGFYAGA
Mb3240|M.bovis_AF2122/97 --------------------------------------------------
Rv3214|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1343|M.marinum_M --------------------------------------------------
MUL_2536|M.ulcerans_Agy99 --------------------------------------------------
MAV_4162|M.avium_104 --------------------------------------------------
MSMEG_1926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0648|M.thermoresistible__bu --------------------------------------------------
Mflv_4679|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1788|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3534|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00808|M.leprae_Br4923 VGWCDARGDGHWVVSIRCAQLSADRRIAFAHAGGGIVAESDPDDEVDETT
Mb3240|M.bovis_AF2122/97 --------------
Rv3214|M.tuberculosis_H37Rv --------------
MMAR_1343|M.marinum_M --------------
MUL_2536|M.ulcerans_Agy99 --------------
MAV_4162|M.avium_104 --------------
MSMEG_1926|M.smegmatis_MC2_155 --------------
TH_0648|M.thermoresistible__bu --------------
Mflv_4679|M.gilvum_PYR-GCK --------------
Mvan_1788|M.vanbaalenii_PYR-1 --------------
MAB_3534|M.abscessus_ATCC_1997 --------------
MLBr_00808|M.leprae_Br4923 AKFATILNALGVKQ