For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAPTVPRQALPCHVGDPDLWFAEAPADLERAKQLCAGCPVRRQCLAAALERAEPWGVWGGEILDRGAVLG FKRPRGRPRKDGRRAGAAA*
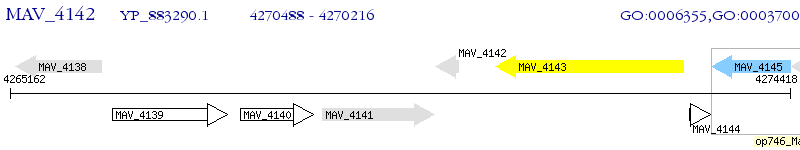
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4142 | - | - | 100% (90) | transcription factor WhiB |
| M. avium 104 | MAV_4223 | - | 4e-09 | 44.90% (49) | transcription factor WhiB |
| M. avium 104 | MAV_4361 | - | 4e-07 | 56.25% (32) | transcription factor WhiB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3221c | whiB7 | 2e-34 | 69.66% (89) | transcriptional regulatory protein WHIB-like WHIB7 |
| M. gilvum PYR-GCK | Mflv_4659 | - | 3e-33 | 65.98% (97) | transcription factor WhiB |
| M. tuberculosis H37Rv | Rv3260c | whiB2 | 2e-08 | 42.86% (49) | transcriptional regulatory protein WHIB-like WHIB2 |
| M. leprae Br4923 | MLBr_00639 | whiB7 | 5e-30 | 66.67% (81) | putative transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3508c | - | 3e-33 | 77.33% (75) | putative transcriptional regulator |
| M. marinum M | MMAR_1365 | whiB7 | 3e-35 | 76.19% (84) | transcriptional regulatory protein Whib-like WhiB7 |
| M. smegmatis MC2 155 | MSMEG_1953 | - | 3e-35 | 77.78% (81) | transcription factor WhiB |
| M. thermoresistible (build 8) | TH_1134 | - | 3e-33 | 75.64% (78) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN WHIB-LIKE WHIB7 |
| M. ulcerans Agy99 | MUL_2511 | whiB7 | 6e-35 | 75.00% (84) | transcriptional regulatory protein Whib-like WhiB7 |
| M. vanbaalenii PYR-1 | Mvan_1809 | - | 7e-34 | 69.23% (91) | transcription factor WhiB |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1365|M.marinum_M -MSMDMSALTVPRQT------LPVLPCHVGNPDLWFAETPADLEQAKELC
MUL_2511|M.ulcerans_Agy99 -----MSALTVPRQT------LPVLPCHVGNPDLWFAETPADLEQAKEFC
MLBr_00639|M.leprae_Br4923 -----MLTLTIPKQT------LPGLPCHADTSDLWFAETPADLECTKTLC
Mb3221c|M.bovis_AF2122/97 -----MSVLTVPRQTP--RQRLPVLPCHVGDPDLWFADTPAGLEVAKTLC
Mflv_4659|M.gilvum_PYR-GCK -----MSSPTCEMTPS-SETRQPAVPCHVEDPDLWFAEDPRDLERAKALC
Mvan_1809|M.vanbaalenii_PYR-1 -----MSSPTCEKT-------KLEVPCHVEDPDLWFAEDPRDLERAKALC
MSMEG_1953|M.smegmatis_MC2_155 -MSIAMTAPTTGVAPMTCETRLPAVPCHVGDPDLWFAENPGDLERAKALC
TH_1134|M.thermoresistible__bu ----------------------VLMPCRTGDPDLWFADDPADLERAKALC
MAB_3508c|M.abscessus_ATCC_199 -----MMTVEVEAR-------RLALPCHVADADLWFAESPADLERAKALC
MAV_4142|M.avium_104 -----MSAPTVPRQ---------ALPCHVGDPDLWFAEAPADLERAKQLC
Rv3260c|M.tuberculosis_H37Rv MVPEAPAPFEEPLPPEATDQWQDRALCAQTDPEAFFPEKGGSTREAKKIC
* .: :*.: . . :* :*
MMAR_1365|M.marinum_M ASCPIRPQCLAAALERAEPWGVWGGEIFERGSIVNRKRPRGRPP-KVAA-
MUL_2511|M.ulcerans_Agy99 ASCPIRPQCLAAALERAEPWGVWGGEIFERGSIVNRKRPRGRPP-KVAA-
MLBr_00639|M.leprae_Br4923 ANCPIRRPCLEAAMERAEPWGVWGGEIFDRGLIVSRKRPRGRPCNDVVVV
Mb3221c|M.bovis_AF2122/97 VSCPIRRQCLAAALQRAEPWGVWGGEIFDQGSIVSHKRPRGRPRKDAVA-
Mflv_4659|M.gilvum_PYR-GCK ADCPLQRACLNAALERQEPWGVWGGEILDRGSVIARKRPRGRPRKDSK--
Mvan_1809|M.vanbaalenii_PYR-1 ADCPLRRECLNAALQRQEPWGVWGGEILERGSVIARKRPRGRPRKDAK--
MSMEG_1953|M.smegmatis_MC2_155 AGCPIRVQCLTAALERQEPWGVWGGEILDRGSIVARKRPRGRPRKDSGG-
TH_1134|M.thermoresistible__bu APCPVRRQCLAAALERQEPWGVWGGEIFDRGSIVARKRPRGRPRKNSD--
MAB_3508c|M.abscessus_ATCC_199 ADCPIRSQCLAAALDRAEPWGVWGGEILEQGTIVARKRPRGRPRKNPLPD
MAV_4142|M.avium_104 AGCPVRRQCLAAALERAEPWGVWGGEILDRGAVLGFKRPRGRPRKDGRR-
Rv3260c|M.tuberculosis_H37Rv MGCEVRHECLEYALAHDERFGIWGG-LSERE---RRRLKRGII-------
* :: ** *: : * :*:*** : :: : **
MMAR_1365|M.marinum_M ------
MUL_2511|M.ulcerans_Agy99 ------
MLBr_00639|M.leprae_Br4923 ------
Mb3221c|M.bovis_AF2122/97 ------
Mflv_4659|M.gilvum_PYR-GCK -SPVAA
Mvan_1809|M.vanbaalenii_PYR-1 -EPVAA
MSMEG_1953|M.smegmatis_MC2_155 -NPAAA
TH_1134|M.thermoresistible__bu --PAAA
MAB_3508c|M.abscessus_ATCC_199 ADTAAA
MAV_4142|M.avium_104 -AGAAA
Rv3260c|M.tuberculosis_H37Rv ------