For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTWNLDGQTANVEQALAGGHTQPAGWFRFYFADQRWEWSEQVQRMHGYEPGSITPTTELVLSHKHPDDRA RVAATIDEIVTNHQAFSTRHRIIDAAGNVRHVVVVGDRLTDEQGEVIGAQGFYIDVSIPHQQVQEEMMSA RLAEISRNRARIEQTKGMLMLIYGISDSAAFELLKWLSQEGNVKLRPLAEQIAEDFRGADVPLPTRSEFD HLLLTAHQRLKASDTRS
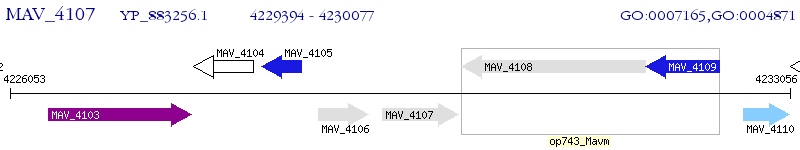
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4107 | - | - | 100% (227) | ANTAR domain-containing protein |
| M. avium 104 | MAV_4562 | - | 3e-70 | 57.01% (221) | ANTAR domain-containing protein |
| M. avium 104 | MAV_3103 | - | 3e-49 | 46.67% (210) | ANTAR domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1090 | - | 4e-64 | 54.63% (216) | putative PAS/PAC sensor protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01796 | - | 3e-33 | 56.20% (121) | hypothetical protein MLBr_01796 |
| M. abscessus ATCC 19977 | MAB_2523c | - | 1e-69 | 58.69% (213) | putative transcription antitermination regulator |
| M. marinum M | MMAR_0736 | - | 2e-54 | 49.75% (199) | putative regulatory protein |
| M. smegmatis MC2 155 | MSMEG_0542 | - | 2e-73 | 59.64% (223) | ANTAR domain-containing protein |
| M. thermoresistible (build 8) | TH_1795 | - | 4e-78 | 64.38% (219) | antar domain protein |
| M. ulcerans Agy99 | MUL_3601 | - | 9e-54 | 49.25% (199) | putative regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_5727 | - | 1e-75 | 62.84% (218) | putative PAS/PAC sensor protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1090|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5727|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2523c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1795|M.thermoresistible__bu --------------------------------------------------
MAV_4107|M.avium_104 --------------------------------------------------
MLBr_01796|M.leprae_Br4923 --------------------------------------------------
MSMEG_0542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0736|M.marinum_M --------------------------------------------------
MUL_3601|M.ulcerans_Agy99 MSSPCHLPTPALSGQVPTVDGLQPSSQRSWCALPRALIGSASTLAQCGAG
Mflv_1090|M.gilvum_PYR-GCK --------------------------------MSTGDSPERTVTGNAP--
Mvan_5727|M.vanbaalenii_PYR-1 --------------------------------MPSDDSVEQALAGGAP--
MAB_2523c|M.abscessus_ATCC_199 -------------------------MSDELSEEEADSELAQALAGGSP--
TH_1795|M.thermoresistible__bu --------------------------------VPGDGSVEDALAGGSP--
MAV_4107|M.avium_104 ---------------------------MTWNLDGQTANVEQALAGGHT--
MLBr_01796|M.leprae_Br4923 --------------------------------------------------
MSMEG_0542|M.smegmatis_MC2_155 --------------------------MTTAPEDDDTAAAVQALTGGTP--
MMAR_0736|M.marinum_M MQVRLGSWWLPGGSGAMISDVQQTAHTGEERPDRGTPSFATPISGAVDSH
MUL_3601|M.ulcerans_Agy99 VQVRLGSWWLPGGSGAMISDVQQTAHTGEERPDRGTPSFATPISGAVDNH
Mflv_1090|M.gilvum_PYR-GCK ARVGWFRYYFADDRWEWSDEVAMLHGYEPGTITPTTEIVLSHRHPDDREE
Mvan_5727|M.vanbaalenii_PYR-1 QRVGWFRFYFADERWEWSEQVQILHGYEPGTVTPTTDLVLSHKHPDDRQQ
MAB_2523c|M.abscessus_ATCC_199 QHVGWFRFYFADERWEWSPQVAQMHGYGPDEITPTTEIILSHKHPDDYRQ
TH_1795|M.thermoresistible__bu QRVGWFRFYFADERWEWSPQVQQMHGYEPGTVTPTTELVLSHKHPDDYAQ
MAV_4107|M.avium_104 QPAGWFRFYFADQRWEWSEQVQRMHGYEPGSITPTTELVLSHKHPDDRAR
MLBr_01796|M.leprae_Br4923 -----------------------MS-------SLTTDLMLTHRHLNDRGQ
MSMEG_0542|M.smegmatis_MC2_155 QQMGWFRFYFADQHWEWSAEVQRMHGYEPGTVTPTTELVLSHKHPDDRDQ
MMAR_0736|M.marinum_M LSIGSFRFWFIGQRWEWSDEVARMHGYEPGAVVPTTKLLLSHKHPDDRAH
MUL_3601|M.ulcerans_Agy99 LSIGSFRFWFIGQRWEWSDEVAGMHGYEPGAVVPTTKLLLSHKHPDDRAH
: **.::*:*:* :* .
Mflv_1090|M.gilvum_PYR-GCK VADMLGQIQHTDDADRAYATRHRIIDTRGRTHEIVIIGDRLLDDTGRTIG
Mvan_5727|M.vanbaalenii_PYR-1 VAATLDEMRRT---HRTFSTRHRIVDTRGDTHDVVVIGDQLLDDAGRLIG
MAB_2523c|M.abscessus_ATCC_199 VAATLQEIRRT---AQAFSTRHRIIDVQQHVHHVVVIGDQLCDDNGVVVG
TH_1795|M.thermoresistible__bu MVATLEQIRRS---HNTFSTRHRIVDKHGDVHSVVVVGDQLCDENGAVIG
MAV_4107|M.avium_104 VAATIDEIVTN---HQAFSTRHRIIDAAGNVRHVVVVGDRLTDEQGEVIG
MLBr_01796|M.leprae_Br4923 VAATIDEILNT---HKLFSTRHRIIDTSENVDNVIVIADQLFDDRGATIG
MSMEG_0542|M.smegmatis_MC2_155 VAVDINDMMTG---RRAFGRRHRIIDTSGTLRQVIVVSDQLHGFDGDVIG
MMAR_0736|M.marinum_M VQDLLDYALRS---GESFSSRHRFVDTAGVVHDAIVVADRMVNESGVVQG
MUL_3601|M.ulcerans_Agy99 VQDLLDYALRS---GESFSSRHRFVDTAGVVHDAIVVADRMVNESGVVQG
: : . :. ***::* :::.*:: . * *
Mflv_1090|M.gilvum_PYR-GCK ARGFCVDMTPSDTQT-KAAISSALAEITENRAAIEQVKGMLMLIYRVDDD
Mvan_5727|M.vanbaalenii_PYR-1 THGFYVDITPTDTQA-KAMVTAALAEITENRAVIEQVKGMLMLIYRVDDE
MAB_2523c|M.abscessus_ATCC_199 THGFYVDVTSSIRQERENSLTVAVVEIAENRSVIEQAKGMLMLVYRLNAD
TH_1795|M.thermoresistible__bu THGFYVDVTPNIAVA-DDLVSAAVAQIAENRATIEQAKGMLMLIYRIGAD
MAV_4107|M.avium_104 AQGFYIDVSIPHQQVQEEMMSARLAEISRNRARIEQTKGMLMLIYGISDS
MLBr_01796|M.leprae_Br4923 TYDFYIDVSALPEQVHEGIVIARLAKSTQNRAGIEQTKSMLMLIDGITYD
MSMEG_0542|M.smegmatis_MC2_155 IRGFYIDVTPPRSRQEDAVITAEVAKITERRAVIEQVKGMLMLIYGIDEN
MMAR_0736|M.marinum_M TSGYYIDLTNTFDETRQEVLDEALPDLFENRAAIEQAKGVLMAVYRVSAE
MUL_3601|M.ulcerans_Agy99 TSGYYIDLTNTFDETRQEVLDEALPDLFENRAAIEQAKGVLMAVYRVSAE
.: :*:: . : : . ..*: ***.*.:** : : .
Mflv_1090|M.gilvum_PYR-GCK TAFELLRWRSQEANVKLRLLAEQLIRDYNELVYDEILPNRATFDHLLLTA
Mvan_5727|M.vanbaalenii_PYR-1 AAFELLRWRSQETNVKLRLLADQLLADYTTLSYDETLPPRATFDHLLLTA
MAB_2523c|M.abscessus_ATCC_199 AAFDVLKWRSQENNVKLRALAQQVVVDFGTLDFDKTLPARSVYDRLLLTA
TH_1795|M.thermoresistible__bu AAFDLLKWRSQETNVKLRMLAEQLIEDFTGLDYQDELPTRGTFDHLLLTA
MAV_4107|M.avium_104 AAFELLKWLSQEGNVKLRPLAEQIAEDFRGA--DVPLPTRSEFDHLLLTA
MLBr_01796|M.leprae_Br4923 TPFNLLKYAVP--------------------------------------G
MSMEG_0542|M.smegmatis_MC2_155 AAFDLLKWLSQENNVKLRVLADQVAKDFTALG-DTGVLSRSKFDQLLLTA
MMAR_0736|M.marinum_M QAFRVLQWRSQETNTKLRALAKQVVAEVASLP-PVSALVQSQFDHLLLTV
MUL_3601|M.ulcerans_Agy99 EAFRVLQWRSQETNTKLRALAKQVVAEVASLP-PVSALVQSQFDHLLLTV
.* :*::
Mflv_1090|M.gilvum_PYR-GCK HQRMSRSGDTVEQ---
Mvan_5727|M.vanbaalenii_PYR-1 HQRVNRAGDTVDG---
MAB_2523c|M.abscessus_ATCC_199 HQRIPR----------
TH_1795|M.thermoresistible__bu HQRTNSDGRRAGIS--
MAV_4107|M.avium_104 HQRLKASDTRS-----
MLBr_01796|M.leprae_Br4923 NQHQAAAG--------
MSMEG_0542|M.smegmatis_MC2_155 ARRAVSSDDDSMSTSA
MMAR_0736|M.marinum_M HERIPAESAS------
MUL_3601|M.ulcerans_Agy99 HERIPAESAS------
.: