For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPRSSIKNEKMYQDLRKKGESKEKAARISNAAAGQGKSSVGRRGGKSGSYQDWTVPELKKRAKELGISGY SGLTKDKLVAKLRNH
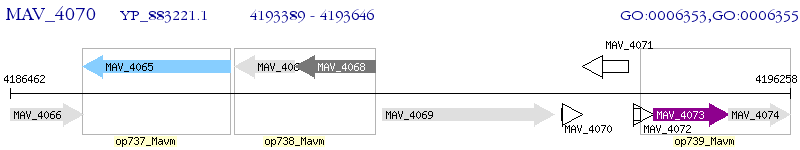
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4070 | - | - | 100% (85) | hypothetical protein MAV_4070 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1968 | - | 2e-35 | 80.00% (85) | hypothetical protein Mflv_1968 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1438 | - | 4e-06 | 36.36% (55) | transcription termination factor Rho |
| M. marinum M | MMAR_4360 | - | 3e-34 | 78.57% (84) | hypothetical protein MMAR_4360 |
| M. smegmatis MC2 155 | MSMEG_1790 | - | 8e-32 | 72.94% (85) | hypothetical protein MSMEG_1790 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0173 | - | 3e-35 | 78.82% (85) | hypothetical protein MUL_0173 |
| M. vanbaalenii PYR-1 | Mvan_4747 | - | 2e-35 | 78.82% (85) | hypothetical protein Mvan_4747 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1968|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4747|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4360|M.marinum_M --------------------------------------------------
MUL_0173|M.ulcerans_Agy99 --------------------------------------------------
MAV_4070|M.avium_104 --------------------------------------------------
MSMEG_1790|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1438|M.abscessus_ATCC_1997 MTDTDLIATEAAPETPEAPAAASSDRAGKERRGDGLGALSTMVLPELRAL
Mflv_1968|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4747|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4360|M.marinum_M --------------------------------------------------
MUL_0173|M.ulcerans_Agy99 --------------------------------------------------
MAV_4070|M.avium_104 --------------------------------------------------
MSMEG_1790|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1438|M.abscessus_ATCC_1997 ASQVGVKGTSGMRKGDLIAAIREHQGALGAPKNNNQAAQSQQPQASSAPA
Mflv_1968|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4747|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4360|M.marinum_M --------------------------------------------------
MUL_0173|M.ulcerans_Agy99 --------------------------------------------------
MAV_4070|M.avium_104 --------------------------------------------------
MSMEG_1790|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1438|M.abscessus_ATCC_1997 EAPQKESGQKESGQKESGLKESAEPTQTALDLPAEAPKPVAESTQDSEGK
Mflv_1968|M.gilvum_PYR-GCK ------MPNSS----IKNEKMYEDLRREGNSKEKAAR-ISNAAAAQGKS-
Mvan_4747|M.vanbaalenii_PYR-1 ------MPNTS----IKNEKMYEELRREGNSKEKAAR-ISNAAAARGKS-
MMAR_4360|M.marinum_M ------MPNPS----IKNEKLYRDLRKEGSSKERAAR-ISNAAAAQGTS-
MUL_0173|M.ulcerans_Agy99 ----MIMPNPS----IKNEKLYRDLRKEGSSKERAAR-ISNAAAAQGTS-
MAV_4070|M.avium_104 ------MPRSS----IKNEKMYQDLRKKGESKEKAAR-ISNAAAGQGKS-
MSMEG_1790|M.smegmatis_MC2_155 ------MPNSS----IKDEKLYQDLRKQGDSKEKAAR-ISNAAASRGRS-
MAB_1438|M.abscessus_ATCC_1997 KAENGAAPREGRGERRRNQNQNQNQPAEGERKQNGSRDNADNAESSGRDG
*. . :::: .: :*. *:..:* :: * . * .
Mflv_1968|M.gilvum_PYR-GCK ----------------SVGRRG--GQS-GSYDDWTVSDLKQRAKQLGMSG
Mvan_4747|M.vanbaalenii_PYR-1 ----------------SVGRKG--GKS-GSYDDWTVGDLKSRAKELGISG
MMAR_4360|M.marinum_M ----------------SVGRKG--GKA-RSYPDWTVPELKKRAKELGMSR
MUL_0173|M.ulcerans_Agy99 ----------------SVGRKG--GKA-RSYPDWTVPELKKRAKELGMSR
MAV_4070|M.avium_104 ----------------SVGRRG--GKS-GSYQDWTVPELKKRAKELGISG
MSMEG_1790|M.smegmatis_MC2_155 ----------------KVGRSG--GKS-GSYEDWTVSDLRSRAKELGITG
MAB_1438|M.abscessus_ATCC_1997 GSSGQNSDNQNQDTNQNQGNRGDDGEGRGRRGRRTRERRRGRDRNNNEGG
. *. * *:. * : * :: .
Mflv_1968|M.gilvum_PYR-GCK YSHLTKGKLID-------KLRNH---------------------------
Mvan_4747|M.vanbaalenii_PYR-1 YSHLSKDKLIS-------KLRNH---------------------------
MMAR_4360|M.marinum_M YSGLTKDELIS-------KLRNR---------------------------
MUL_0173|M.ulcerans_Agy99 YSGLTKDELIS-------KLRNH---------------------------
MAV_4070|M.avium_104 YSGLTKDKLVA-------KLRNH---------------------------
MSMEG_1790|M.smegmatis_MC2_155 YSDKNKGELVK-------MLRNH---------------------------
MAB_1438|M.abscessus_ATCC_1997 ETELREDDVVQPVAGILDVLDNYAFVRTSGYLAGPNDVYVSMNLVRKNGL
: :..:: * *
Mflv_1968|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4747|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4360|M.marinum_M --------------------------------------------------
MUL_0173|M.ulcerans_Agy99 --------------------------------------------------
MAV_4070|M.avium_104 --------------------------------------------------
MSMEG_1790|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1438|M.abscessus_ATCC_1997 RRGDAITGAVRVPRDPDAGNQNQRQKFNPLVRLDSVNGGPVEDAKKRPDF
Mflv_1968|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4747|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4360|M.marinum_M --------------------------------------------------
MUL_0173|M.ulcerans_Agy99 --------------------------------------------------
MAV_4070|M.avium_104 --------------------------------------------------
MSMEG_1790|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1438|M.abscessus_ATCC_1997 TKLTPLYPNQRLRLETTTEKLTTRVIDLIMPIGKGQRALIVSPPKAGKTT
Mflv_1968|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4747|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4360|M.marinum_M --------------------------------------------------
MUL_0173|M.ulcerans_Agy99 --------------------------------------------------
MAV_4070|M.avium_104 --------------------------------------------------
MSMEG_1790|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1438|M.abscessus_ATCC_1997 IMQNIANAISANNPECHLMVVLVDERPEEVTDMQRSVKGEVIASTFDRPP
Mflv_1968|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4747|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4360|M.marinum_M --------------------------------------------------
MUL_0173|M.ulcerans_Agy99 --------------------------------------------------
MAV_4070|M.avium_104 --------------------------------------------------
MSMEG_1790|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1438|M.abscessus_ATCC_1997 SDHTQAAELAIERAKRLVEQGKDVVVLLDSITRLGRAYNNASPASGRILS
Mflv_1968|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4747|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4360|M.marinum_M --------------------------------------------------
MUL_0173|M.ulcerans_Agy99 --------------------------------------------------
MAV_4070|M.avium_104 --------------------------------------------------
MSMEG_1790|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1438|M.abscessus_ATCC_1997 GGVDSTALYPPKRFLGAARNIENGGSLTIIATAMVETGSTGDTVIFEEFK
Mflv_1968|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4747|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4360|M.marinum_M --------------------------------------------------
MUL_0173|M.ulcerans_Agy99 --------------------------------------------------
MAV_4070|M.avium_104 --------------------------------------------------
MSMEG_1790|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1438|M.abscessus_ATCC_1997 GTGNAELKLDRKIAERRVFPAVDVNPSGTRKDELLMSGDEFAIVHKLRRV
Mflv_1968|M.gilvum_PYR-GCK ----------------------------------------------
Mvan_4747|M.vanbaalenii_PYR-1 ----------------------------------------------
MMAR_4360|M.marinum_M ----------------------------------------------
MUL_0173|M.ulcerans_Agy99 ----------------------------------------------
MAV_4070|M.avium_104 ----------------------------------------------
MSMEG_1790|M.smegmatis_MC2_155 ----------------------------------------------
MAB_1438|M.abscessus_ATCC_1997 LSGLDSHQAIDLLMSQLRKTKNNYEFLVQVSKNTPGAAGAEETQYS