For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PARTALSPGELSPTLPVPARIPRPEYVGKPTAREGSEPWVQTPEVIEKMRVAGRIAAGALVEAGKAVAPG VTTDELDRIAHEYMIDHGAYPSTLGYKGYPKSCCTSLNEVICHGIPDSTVIEDGDIVNIDVTAYIDGVHG DTNATFLAGDVAEEHRLLVERTREATMRAINAVKPGRALSVIGRVIESYANRFGYNVVRDFTGHGIGTTF HNGLVVLHYDQPSVSTIMQPGMTFTIEPMINLGGLDYEIWDDGWTVVTKDRKWTAQFEHTLLVTDTGVEI LTLP*
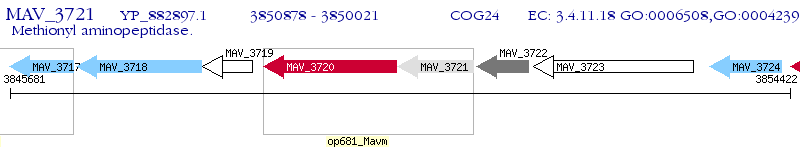
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3721 | map | - | 100% (285) | methionine aminopeptidase |
| M. avium 104 | MAV_4432 | map | 2e-41 | 38.78% (245) | methionine aminopeptidase |
| M. avium 104 | MAV_3412 | - | 3e-11 | 29.15% (199) | peptidase, M24 family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2886c | mapB | 1e-154 | 92.23% (283) | methionine aminopeptidase |
| M. gilvum PYR-GCK | Mflv_4070 | - | 1e-149 | 87.99% (283) | methionine aminopeptidase |
| M. tuberculosis H37Rv | Rv2861c | mapB | 1e-154 | 92.23% (283) | methionine aminopeptidase |
| M. leprae Br4923 | MLBr_01576 | mapB | 1e-157 | 92.96% (284) | methionine aminopeptidase |
| M. abscessus ATCC 19977 | MAB_3164c | - | 1e-129 | 81.89% (265) | methionine aminopeptidase (MAP) |
| M. marinum M | MMAR_1842 | mapB | 1e-154 | 92.28% (285) | methionine aminopeptidase MapB |
| M. smegmatis MC2 155 | MSMEG_2587 | map | 1e-150 | 88.34% (283) | methionine aminopeptidase |
| M. thermoresistible (build 8) | TH_1393 | map | 1e-149 | 87.99% (283) | PROBABLE METHIONINE AMINOPEPTIDASE MAPB (MAP) (PEPTIDASE M) |
| M. ulcerans Agy99 | MUL_2091 | mapB | 1e-147 | 93.28% (268) | methionine aminopeptidase MapB |
| M. vanbaalenii PYR-1 | Mvan_2272 | - | 1e-150 | 87.63% (283) | methionine aminopeptidase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_3721|M.avium_104 MPARTALSPGELSPTLPVPARIPRPEYVGKPTAREGS-EPWVQTPEVIEK
MLBr_01576|M.leprae_Br4923 MPARTALSPGFLSPTLPVPAWIPRPEYVGKPTAQEGS-ESWVQTPEVVEK
Mb2886c|M.bovis_AF2122/97 MPSRTALSPGVLSPTRPVPNWIARPEYVGKPAAQEGS-EPWVQTPEVIEK
Rv2861c|M.tuberculosis_H37Rv MPSRTALSPGVLSPTRPVPNWIARPEYVGKPAAQEGS-EPWVQTPEVIEK
MMAR_1842|M.marinum_M MPARTALSPGVLSPMRSVPKWIARPEYVGKPTAREGT-EPWVQTPEVIEK
MUL_2091|M.ulcerans_Agy99 --------------MRSVPKWIARPEYVGKPTAREGT-EPWVQTPEVIEK
Mflv_4070|M.gilvum_PYR-GCK MSVRTALRPGTVSPMLAVPKSIERPEYVGKPTAREGS-EPWVQTPEVIEK
Mvan_2272|M.vanbaalenii_PYR-1 MSVRTALRPGTVSPMLPVPKSIVRPEYVGRPTAREGN-EPWVQTPEVIEK
MSMEG_2587|M.smegmatis_MC2_155 MSVRSALRPGVLSPTLPVPKSIPRPEYAWKPTVQEGS-EPWVQTPEVIEK
TH_1393|M.thermoresistible__bu MSVRTALRPGVISPTRPVPKSIERPEYVWKPTANEGN-EPWVQTPEVIDK
MAB_3164c|M.abscessus_ATCC_199 -------------------MSIDRPEYVGKSKVAEAIGEPYVQTPEVIEK
* ****. :. . *. *.:******::*
MAV_3721|M.avium_104 MRVAGRIAAGALVEAGKAVAPGVTTDELDRIAHEYMIDHGAYPSTLGYKG
MLBr_01576|M.leprae_Br4923 MRVAGRIAAGALTEAGKAVAPGVTTDELDRIVHEYMVNHGAYPSTLGYKG
Mb2886c|M.bovis_AF2122/97 MRVAGRIAAGALAEAGKAVAPGVTTDELDRIAHEYLVDNGAYPSTLGYKG
Rv2861c|M.tuberculosis_H37Rv MRVAGRIAAGALAEAGKAVAPGVTTDELDRIAHEYLVDNGAYPSTLGYKG
MMAR_1842|M.marinum_M MRVAGRIAAGALVEAGKAVAPGVTTDELDRIAHEYMVDNGAYPSTLEYKG
MUL_2091|M.ulcerans_Agy99 MRVAGRIAAGALVEAGKAVAPGVTTDELDRIAHEYMVDNGAYPSTLEYKG
Mflv_4070|M.gilvum_PYR-GCK MRVAGRIAAGALAEAGKAVAPGVTTDRLDRIAHDYMVDHGAYPSTLGYKG
Mvan_2272|M.vanbaalenii_PYR-1 MRVAGRIAAGALAEAGKAVEPGVTTDQLDRIAHEYMVDHGAYPSTLGYKG
MSMEG_2587|M.smegmatis_MC2_155 MRVAGRIAAGALAEAGRAVAPGVTTDELDRIAHEYMIDHGAYPSTLGYKG
TH_1393|M.thermoresistible__bu MRIAGRIAAGALAEAGKAVEPGVTTDELDRIVHEYLIDHGAYPSTLGYKG
MAB_3164c|M.abscessus_ATCC_199 MRVAGRIAARALALAGEAVAPGVTTDELDRIAHDYMVSQGAYPSTLGYKG
**:****** **. **.** ******.****.*:*::.:******* ***
MAV_3721|M.avium_104 YPKSCCTSLNEVICHGIPDSTVIEDGDIVNIDVTAYIDGVHGDTNATFLA
MLBr_01576|M.leprae_Br4923 YPKSCCTSLNEVICHGIPDSTVIADGDIVNIDVTAYIDGVHGDTNATFLS
Mb2886c|M.bovis_AF2122/97 FPKSCCTSLNEVICHGIPDSTVITDGDIVNIDVTAYIGGVHGDTNATFPA
Rv2861c|M.tuberculosis_H37Rv FPKSCCTSLNEVICHGIPDSTVITDGDIVNIDVTAYIGGVHGDTNATFPA
MMAR_1842|M.marinum_M FPKSCCTSLNEVICHGIPDSTVIADGDIVNIDVTAYFDGVHGDTNATFLA
MUL_2091|M.ulcerans_Agy99 FPKSCCTSLNEVICHGIPDSTVIADGDIVNIDVTAYFDGVHGDTNATFLA
Mflv_4070|M.gilvum_PYR-GCK FPKSCCTSLNEVICHGIPDSTVIEDGDIVNIDVTAYIDGVHGDTNATFLA
Mvan_2272|M.vanbaalenii_PYR-1 FPKSCCTSLNEVICHGIPDDTVVEDGDIVNIDVTAYIDGVHGDTNATFLA
MSMEG_2587|M.smegmatis_MC2_155 FPKSCCTSLNEIICHGIPDSTVIEDGDIVNIDVTAYIDGVHGDTNATFLA
TH_1393|M.thermoresistible__bu FPKSCCTSLNEVICHGIPDSTVIEDGDIVNIDVTAYIHGVHGDTNATFLA
MAB_3164c|M.abscessus_ATCC_199 FPKSCCTSLNEIICHGIPDSTVIEDGDIVNIDVTAYIDGVHGDTNATFLA
:**********:*******.**: ************: ********** :
MAV_3721|M.avium_104 GDVAEEHRLLVERTREATMRAINAVKPGRALSVIGRVIESYANRFGYNVV
MLBr_01576|M.leprae_Br4923 GDVSESHRLLVERTREATMRAINAVKPGRALSIIGRVIESYANRFGYNVV
Mb2886c|M.bovis_AF2122/97 GDVADEHRLLVDRTREATMRAINTVKPGRALSVIGRVIESYANRFGYNVV
Rv2861c|M.tuberculosis_H37Rv GDVADEHRLLVDRTREATMRAINTVKPGRALSVIGRVIESYANRFGYNVV
MMAR_1842|M.marinum_M GDVSEEHRLLVERTREATMRAINAVKPGRALSVIGRVIEAYANRFGYNVV
MUL_2091|M.ulcerans_Agy99 GDVSEEHRLLVERTREATMRAINAVKPGRALSVIGRVIEAYANRFGYNVV
Mflv_4070|M.gilvum_PYR-GCK GNVSEEHRLLVERTHEATMRAIKAVKPGRALSVVGRVIEAYANRFGYNVV
Mvan_2272|M.vanbaalenii_PYR-1 GDVSEEHRLLVERTHEATMRAIKAVKPGRALSIVGRVIESYANRFGYNVV
MSMEG_2587|M.smegmatis_MC2_155 GDVSEEHRLLVERTHEATMRAIKAVKPGRALSVVGRVIEAYANRFGYNVV
TH_1393|M.thermoresistible__bu GDVSEEHRLLVERTREAMMRAIKAVKPGRALSVVGRVIESYANRFGYNVV
MAB_3164c|M.abscessus_ATCC_199 GDVSEEHRLLVERTREATMRAINAVKPGRALSVVGRVIEAYAKRFGYNVV
*:*::.*****:**:** ****::********::*****:**:*******
MAV_3721|M.avium_104 RDFTGHGIGTTFHNGLVVLHYDQPSVSTIMQPGMTFTIEPMINLGGLDYE
MLBr_01576|M.leprae_Br4923 RHFTGHGIGTTFHNGLVVLHYDQPFVTTIMQPGMTFTIEPMINLGTLDYE
Mb2886c|M.bovis_AF2122/97 RDFTGHGIGTTFHNGLVVLHYDQPAVETIMQPGMTFTIEPMINLGALDYE
Rv2861c|M.tuberculosis_H37Rv RDFTGHGIGTTFHNGLVVLHYDQPAVETIMQPGMTFTIEPMINLGALDYE
MMAR_1842|M.marinum_M RDFTGHGIGTTFHNGLVVLHYDQPAVETIMQTGMTFTIEPMINLGTLDYE
MUL_2091|M.ulcerans_Agy99 RDFTGHGIGTTFHNGLVVLHYDQPAVETIMQTGMTFTIEPMINLGTLDYE
Mflv_4070|M.gilvum_PYR-GCK RDFTGHGIGTTFHNGLVVLHYDQPSVQTVLEPGMTFTIEPMINLGSLDYE
Mvan_2272|M.vanbaalenii_PYR-1 RDFTGHGIGTTFHNGLVVLHYDQPAVETMLEPGMTFTIEPMINVGGLDYE
MSMEG_2587|M.smegmatis_MC2_155 RDFTGHGIGTTFHNGLVVLHYDQPAVETVLEPGMTFTIEPMINLGTLDYE
TH_1393|M.thermoresistible__bu RDFTGHGIGTTFHNGLVVLHYDEPSVTTVMEPGMTFTIEPMINLGSLEYE
MAB_3164c|M.abscessus_ATCC_199 RAFTGHGVGPTFHNGLIVPHYDDPNLDIVIEPGMTFTIEPMINLGSLEYE
* *****:*.******:* ***:* : :::.***********:* *:**
MAV_3721|M.avium_104 IWDDGWTVVTKDRKWTAQFEHTLLVTDTGVEILTLP
MLBr_01576|M.leprae_Br4923 VWDDGWTVVTKDRKWTAQFEHTLLVTDTGVEILTVQ
Mb2886c|M.bovis_AF2122/97 IWDDGWTVVTKDRKWTAQFEHTLLVTDTGVEILTCL
Rv2861c|M.tuberculosis_H37Rv IWDDGWTVVTKDRKWTAQFEHTLLVTDTGVEILTCL
MMAR_1842|M.marinum_M IWDDDWTVVTKDRKWTAQFEHTLLVTDTGAEILTLP
MUL_2091|M.ulcerans_Agy99 IWDDDWTVVTKDRKWTAQFEHTLLVTDTGAEILTLP
Mflv_4070|M.gilvum_PYR-GCK IWDDDWTVATRDKKWTAQFEHTLVVTESGAEILTQL
Mvan_2272|M.vanbaalenii_PYR-1 IWDDDWTVVTKDKKWTAQFEHTLVVTESGAEILTQL
MSMEG_2587|M.smegmatis_MC2_155 IWDDGWTVATTDGKWTAQFEHTLVVTEDGAEILTQL
TH_1393|M.thermoresistible__bu IWDDGWTVVTKDRKWTAQFEHTLVVTEDGAEILTQL
MAB_3164c|M.abscessus_ATCC_199 IWDDTWTVATADKLWTAQFEHTMVVTEDGVEILTVP
:*** ***.* * ********::**: *.****