For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KPYPFSAIVGHDRLRLALLLCAVRPEIGGALIRGEKGTAKSTAVRGLAALLSAATGSDANLVEMPLGATE DRVIGSLDLQRVLRDGEHAFSPGLLARAHGGVLYVDEVNLLHDHLVDVLLDAAAMGRVHIERDGISHSHD ARFVLIGTMNPEEGELRPQLLDRFGLTVDVHASRDVEVRAQVIRQRMAYEADPDGFAERYADADAELARR IAAARALVDDVVLPDNELRRIAALCAAFDVDGMRADLVVARTAVAHAAWRGAPTVEEQDIRVAAELALPH RRRRDPFDDPGIDREQLDEALAQAGSEPDPDPDPPGGGQAGQTEQQPPQRDSAQDAKPRPPTTKPSAPPS KTFRTRALRVPGVGEGAPGRRSRARNASGSVIAATDGADTGCGGHGLHLFATLLSAAERAGAGPLRPGPD DVRRAVREGREGNLVIFVVDASGSMAARDRMAAVSGATLSLLRDAYQRRDKVAVITFRQQQARLLLPPTS SAHIAGRRLARFDTGGKTPLAEGLLAARELIVREKARDRARRALVVVLTDGRATAGPDPLGRSRIAAARL VAEGAAAVVVDCETSYVRLGLAEHLARQLGAPAVRLEQLHADSLTRAVRDAA*
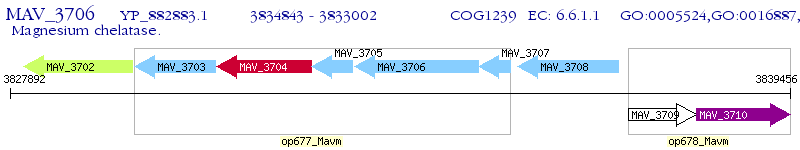
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3706 | - | - | 100% (613) | chelatase |
| M. avium 104 | MAV_4052 | - | 1e-07 | 26.32% (285) | ATPase family protein associated with various cellular |
| M. avium 104 | MAV_3751 | - | 2e-05 | 26.59% (267) | Mg-chelatase subunits D/I family protein, ComM subfamily |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2875c | - | 0.0 | 83.52% (631) | magnesium chelatase |
| M. gilvum PYR-GCK | Mflv_4050 | - | 0.0 | 78.81% (623) | magnesium chelatase |
| M. tuberculosis H37Rv | Rv2850c | - | 0.0 | 83.84% (631) | magnesium chelatase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3157c | - | 0.0 | 70.92% (619) | magnesium-chelatase |
| M. marinum M | MMAR_1883 | - | 0.0 | 83.52% (619) | magnesium chelatase |
| M. smegmatis MC2 155 | MSMEG_2615 | - | 0.0 | 76.52% (626) | chelatase |
| M. thermoresistible (build 8) | TH_1386 | - | 1e-117 | 86.78% (242) | POSSIBLE MAGNESIUM CHELATASE |
| M. ulcerans Agy99 | MUL_2111 | - | 0.0 | 83.36% (619) | magnesium chelatase |
| M. vanbaalenii PYR-1 | Mvan_2302 | - | 0.0 | 76.61% (637) | magnesium chelatase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2875c|M.bovis_AF2122/97 MKP-------YPFSAIVGHDRLRLALLLCAVRPEIGGALIRGEKGTAKST
Rv2850c|M.tuberculosis_H37Rv MKP-------YPFSAIVGHDRLRLALLLCAVRPEIGGALIRGEKGTAKST
MMAR_1883|M.marinum_M MKA-------YPFSAIVGHDQMRLALLLCAVRPEIGGVLIRGEKGTAKST
MUL_2111|M.ulcerans_Agy99 MKA-------YPFSAIVGHGPMRLALLLCAVRPEIGGVLIRGEKGTAKST
MAV_3706|M.avium_104 MKP-------YPFSAIVGHDRLRLALLLCAVRPEIGGALIRGEKGTAKST
Mflv_4050|M.gilvum_PYR-GCK MTG-TASPPIYPLSAVIGHDRLRLALVLCAVRPDIGGVLIRGEKGTAKST
Mvan_2302|M.vanbaalenii_PYR-1 MTSPESSPPIYPFSAVIGHDRLRLALVLCAVRPDIGGVLIRGEKGTAKST
MSMEG_2615|M.smegmatis_MC2_155 MIQQT-----YPLSAIVGQDRLRLALVLCAVRPEIGGVLIRGEKGTAKST
MAB_3157c|M.abscessus_ATCC_199 MASATAG---YPLSAIVGHDRLRLALVLSAVRPDIGGVLIRGEKGTAKST
TH_1386|M.thermoresistible__bu VITTT-----YPFSAIVGHDRLRLALLLCAVRPEIGGVLIRGEKGTAKST
: **:**::*:. :****:*.****:***.************
Mb2875c|M.bovis_AF2122/97 AVRGLAALLS---VATGSTETGLVELPLGATEDRVVGSLDLQRVMRDGEH
Rv2850c|M.tuberculosis_H37Rv AVRGLAALLS---VATGSTETGLVELPLGATEDRVVGSLDLQRVMRDGEH
MMAR_1883|M.marinum_M AVRGLAALLS---AATGSSGPGLVEMPLGATEDRVVGSLDLQRVLRDGEH
MUL_2111|M.ulcerans_Agy99 AVRGLAALLS---AATGSSGPGLVEMPLGATEDRVVGSLDLQRVLRDGEH
MAV_3706|M.avium_104 AVRGLAALLS---AATG-SDANLVEMPLGATEDRVIGSLDLQRVLRDGEH
Mflv_4050|M.gilvum_PYR-GCK AVRGLAQVLSAAFAAAGDDGGQLVELPIGATEDRVVGSLDLQKVLRDGEH
Mvan_2302|M.vanbaalenii_PYR-1 AVRGLAHVLSAAFAASGEHGGQLVELPIGATEDRVVGSLDLQKVLRDGEH
MSMEG_2615|M.smegmatis_MC2_155 AVRGLAAVLA-----AVDEDAKLVELPIGATEDRVVGSLDLQKVLRDGEH
MAB_3157c|M.abscessus_ATCC_199 AVRALASVLG------SVDGARLVELPIGATEDRVVGSLDLQKVLRDGEH
TH_1386|M.thermoresistible__bu AVRGLAHILH------AVDGASLVELPIGATEDRVVGSLDLQKVLRDGEH
***.** :* ***:*:*******:******:*:*****
Mb2875c|M.bovis_AF2122/97 AFSPGLLARAHGGVLYVDEVNLLHDHLVDILLDAAAMGRVHVERDGISHS
Rv2850c|M.tuberculosis_H37Rv AFSPGLLARAHGGVLYVDEVNLLHDHLVDILLDAAAMGRVHVERDGISHS
MMAR_1883|M.marinum_M AFAPGLLARAHGGVLYVDEVNLLHDHLVDILLDAAAMGRVHVERDGISHS
MUL_2111|M.ulcerans_Agy99 AFAPGLLARAHGGVLYVDEVNLLHDHLVDILLDAAAMGRVHVERDGISHS
MAV_3706|M.avium_104 AFSPGLLARAHGGVLYVDEVNLLHDHLVDVLLDAAAMGRVHIERDGISHS
Mflv_4050|M.gilvum_PYR-GCK AFSPGLLARAHRGVLYVDEVNLLHDHLVDVLLDAAAMGRVHIERDGVSHS
Mvan_2302|M.vanbaalenii_PYR-1 AFSPGLLARAHRGVLYVDEVNLLHDHLVDVLLDAAAMGRVHIERDGVSHS
MSMEG_2615|M.smegmatis_MC2_155 AFSPGLLARAHGGVLYVDEVNLLHDHLVDVLLDAAAMGRVHVERDGISHS
MAB_3157c|M.abscessus_ATCC_199 AFSPGLLARADGGVLYVDEVNLLHDHLVDVVLDAAAMGRVHVERDGVSHS
TH_1386|M.thermoresistible__bu AFSPGLLARAHGGVLYVDEVNLLHDHLVDVLLDAAAMGRVHVERDGVSHS
**:*******. *****************::**********:****:***
Mb2875c|M.bovis_AF2122/97 HEARFVLIGTMNPEEGELRPQLLDRFGLTVDVQASRDIDVRVQVIRRRMA
Rv2850c|M.tuberculosis_H37Rv HEARFVLIGTMNPEEGELRPQLLDRFGLTVDVQASRDIDVRVQVIRRRMA
MMAR_1883|M.marinum_M HEARFVLIGTMNPEEGELRPQLLDRFGLTVDVHASRDVDVRVEVIRQRMA
MUL_2111|M.ulcerans_Agy99 HEARFVLIGTMNPEEGELRPQLLDRFGLTVDVHASRDVDVRVEVIRQRMA
MAV_3706|M.avium_104 HDARFVLIGTMNPEEGELRPQLLDRFGLTVDVHASRDVEVRAQVIRQRMA
Mflv_4050|M.gilvum_PYR-GCK HDARFVLIGTMNPEEGELRPQLLDRFGLTVDVAASRDVDVRAEVIRTRLA
Mvan_2302|M.vanbaalenii_PYR-1 HDARFVLIGTMNPEEGELRPQLLDRFGLTVDVAASRDVDVRVEVIRSRLA
MSMEG_2615|M.smegmatis_MC2_155 HEARFVLIGTMNPEEGELRPQLLDRFGLTVDVTASRDVDVRVEVIRARMD
MAB_3157c|M.abscessus_ATCC_199 YDARFVLIGTMNPEEGELRPQLLDRFGFAVDIHASRDVEVRAEVIRRRLA
TH_1386|M.thermoresistible__bu HEARFVLIGTMNPEEGELRPQLLDRFGLTVDVVASRDIDVRTEVVRQRMA
::*************************::**: ****::**.:*:* *:
Mb2875c|M.bovis_AF2122/97 YEADPDAFVARYADADAELAHRIAAARATVDDVVLGDNELRRIAALCAAF
Rv2850c|M.tuberculosis_H37Rv YEADPDAFVARYADADAELAHRIAAARATVDDVVLGDNELRRIAALCAAF
MMAR_1883|M.marinum_M FEADPDAFCERYVGPDAELARRIAEARDIVDTVVLPDHELRRIAALCAAF
MUL_2111|M.ulcerans_Agy99 FEADPDAFCERYVGPDAELARRIAEARDIVDTVVLPDHELRRIAALCAAF
MAV_3706|M.avium_104 YEADPDGFAERYADADAELARRIAAARALVDDVVLPDNELRRIAALCAAF
Mflv_4050|M.gilvum_PYR-GCK YEADPVAFARRYADADADLAARIADARALVPDVVLPDRELRRIAALCAAF
Mvan_2302|M.vanbaalenii_PYR-1 YEADPAAFAERYAAADADLAAQIAKARALIADVVLPDNELRRIAALCAAF
MSMEG_2615|M.smegmatis_MC2_155 FEADPEGFVRRHAEADAELSRRIAAARDLVDSVVLPDTELRRIAALCAAF
MAB_3157c|M.abscessus_ATCC_199 YEADPAGFVSRYASEEAELAARIADARQLLARVVLPDAELRRIATLCAAF
TH_1386|M.thermoresistible__bu FEADPEGFAARYRDADAGLARRVAAARALVSSVELPDKELRRIAALCAAF
:**** .* *: :* *: ::* ** : * * * ******:*****
Mb2875c|M.bovis_AF2122/97 DVDGMRADLVVARTAAAHAAWR----------GVRTVEEQDIQAAAELAL
Rv2850c|M.tuberculosis_H37Rv DVDGMRADLVVARTAAAHAAWR----------GVRTVEEQDIRAAAELAL
MMAR_1883|M.marinum_M DVDGMRADLVLARTATAHAAWR----------GAPAVEEQDVRVAAELAL
MUL_2111|M.ulcerans_Agy99 DVDGMRADLVLARTATAHAAWR----------GAPAVEEQDVRVAAELAL
MAV_3706|M.avium_104 DVDGMRADLVVARTAVAHAAWR----------GAPTVEEQDIRVAAELAL
Mflv_4050|M.gilvum_PYR-GCK DVDGMRADLVVARTAMAHAAWR----------GEVTVDEEDIRVAAELAL
Mvan_2302|M.vanbaalenii_PYR-1 DVDGMRADLVVARTAVAHAAWRVSATGAGEATGDVTVGEEDIRVAAELAL
MSMEG_2615|M.smegmatis_MC2_155 DVDGMRADLVVARTAVAHAAWR----------GAGTVSEEDVRVAAELAL
MAB_3157c|M.abscessus_ATCC_199 DVDGMRADLVVARAAIAHAAWR----------GADTVGEPDIRVAAELAL
TH_1386|M.thermoresistible__bu DVDGMRX-----------AARR----------------------------
****** ** *
Mb2875c|M.bovis_AF2122/97 PHRRRRDPFDDHGIDRDQLDEALALASVD------------PEPEP----
Rv2850c|M.tuberculosis_H37Rv PHRRRRDPFDDHGIDRDQLDEALALASVD------------PEPEP----
MMAR_1883|M.marinum_M PHRRRRDPFDDHGIDRDQLDEALARAGAESQGS--------PEPEP----
MUL_2111|M.ulcerans_Agy99 PHRRRRDPFDDHGIDRDQLDEALARAGAESQGS--------PEPEP----
MAV_3706|M.avium_104 PHRRRRDPFDDPGIDREQLDEALAQAGSE------------PDPDP----
Mflv_4050|M.gilvum_PYR-GCK PHRRRRDPFDDPGLDPGTLDEAMEQAGESAGPTP------DPEPDP----
Mvan_2302|M.vanbaalenii_PYR-1 PHRRRRDPFDDPGLDPDRLDEAMEQAGESVDGPP------DPDPEP----
MSMEG_2615|M.smegmatis_MC2_155 PHRRRRDPFDDPGLDPDRLDEAMQQAGEQAEAGPESNPETDPETDPDFDP
MAB_3157c|M.abscessus_ATCC_199 PHRRRRDPFDEPGLDPDQLDQAMRDSAPPQDQDGE-----DPDPEP----
TH_1386|M.thermoresistible__bu PPRR----------------------------------------------
* **
Mb2875c|M.bovis_AF2122/97 --DPPGGGQSANEPASQPNSRSKSTEPGAPSSMGDDPPRPASPRLRSSPR
Rv2850c|M.tuberculosis_H37Rv --DPPGGGQSANEPASQPNSRSKSTEPGAPSSMGDDPPRPASPRLRSSPR
MMAR_1883|M.marinum_M --DPPGGGQSAGQDCPPQQQNSNS-------------------RPQQAPK
MUL_2111|M.ulcerans_Agy99 --DPPGGGQSAGQDCPPQQQNSNS-------------------RPQQAPK
MAV_3706|M.avium_104 --DPPGGGQAGQTEQQPPQRDSAQDAK----------------PRPPTTK
Mflv_4050|M.gilvum_PYR-GCK --DPPGGGADADGSADGQPPQQNSSSH-----------------PSSSTR
Mvan_2302|M.vanbaalenii_PYR-1 --DPPGGGSNPDDVAEGQERQQKSS---------------------SPSG
MSMEG_2615|M.smegmatis_MC2_155 DFDPPGGGATTDSDAAAPQGKPKS----------------------SSTR
MAB_3157c|M.abscessus_ATCC_199 --DGPGGGASDSAPDPAKASDSQQDS----------------AYAAGSSR
TH_1386|M.thermoresistible__bu ---PPGRCRAPTAS--------------------------------SDRK
**
Mb2875c|M.bovis_AF2122/97 PSAPPSKIFRTRALRVPGVGTGAPGRRSRARNASGSVVAAAEVSDPD--A
Rv2850c|M.tuberculosis_H37Rv PSAPPSKIFRTRALRVPGVGTGAPGRRSRARNASGSVVAAAEVSDPD--A
MMAR_1883|M.marinum_M PSAGPSKVFRTRALVVPGVGEGAPGRRSRARNGSGSVVAAADGSDSNSGA
MUL_2111|M.ulcerans_Agy99 PSAGPSKVFRTRALVVPGVGEGAPGRRSRARNGSGSVVAAADGSDSNSGA
MAV_3706|M.avium_104 PSAPPSKTFRTRALRVPGVGEGAPGRRSRARNASGSVIAATDGADTGCGG
Mflv_4050|M.gilvum_PYR-GCK QSAPPSPTFKTRTLVVPGVGEGAPGRRSRARNRTGSVVSATSDPHVG---
Mvan_2302|M.vanbaalenii_PYR-1 QSAPPAHTFKTRALVVPGVGEGAPGRRSRARNRTGTVIAATEDAGAG---
MSMEG_2615|M.smegmatis_MC2_155 QSAPPAATFRTRALVVPGIGEGAPGKRSRARNRTGKPIAATAEPGEG---
MAB_3157c|M.abscessus_ATCC_199 PSPAPSATFRTKTLRVPGVGMGAPGKRSVARNRAGKVIAPSSDEGFG---
TH_1386|M.thermoresistible__bu SSRGWPESLRELSCRFS---------------------------------
* . :: : ..
Mb2875c|M.bovis_AF2122/97 HGLHLFATLLAAGERAFGAGPL-RPWPDDVRRAIRESREGNLVIFVVDAS
Rv2850c|M.tuberculosis_H37Rv HGLHLFATLLAAGERAFGAGPL-RPWPDDVRRAIREGREGNLVIFVVDAS
MMAR_1883|M.marinum_M RGLHLFATLLSAAENATGSGPL-RPQGADIRRAVREGREGNLVIFVVDAS
MUL_2111|M.ulcerans_Agy99 RGLHLFATLLSAAENATGSGPL-RPQGADIRRAVREGREGNLVIFVVDAS
MAV_3706|M.avium_104 HGLHLFATLLSAAERA-GAGPL-RPGPDDVRRAVREGREGNLVIFVVDAS
Mflv_4050|M.gilvum_PYR-GCK HGMHVFATLLAAAGRQTRSGAV-RPGPDDVHRAVREGREGNLVIFVVDAS
Mvan_2302|M.vanbaalenii_PYR-1 HGLHMFATLLNAAGRRRGPGAV-RPGPDDVRRAVREGREGNLVIFVVDAS
MSMEG_2615|M.smegmatis_MC2_155 HGVHVFGTLLATAGRQREPGRP-RPVPEDIRRAVREGREGNLVIFVVDAS
MAB_3157c|M.abscessus_ATCC_199 --VHVIGTLMSAASRVTEPGRLPRPVLTDLQWAIREGREGNLVIFVVDAS
TH_1386|M.thermoresistible__bu --IHRMMFLPNGFEQATTAGLG------DVDVAVNLGVDP----------
:* : * . .* *: *:. . :
Mb2875c|M.bovis_AF2122/97 GSMAARDRMAAVSGATLSLLRDAYQRRDKVAVITFRQHEATLLLSPTSSA
Rv2850c|M.tuberculosis_H37Rv GSMAARDRMAAVSGATLSLLRDAYQRRDKVAVITFRQHEATLLLSPTSSA
MMAR_1883|M.marinum_M GSMAARDRMAAVSGATMSLLRDAYQRRDKVAVITFRQHEAKILLSPTSSA
MUL_2111|M.ulcerans_Agy99 GSMAARDRMAAVSGATMSLLRDAYQRRDKVAVITFRQHEAKILLSPTSSA
MAV_3706|M.avium_104 GSMAARDRMAAVSGATLSLLRDAYQRRDKVAVITFRQQQARLLLPPTSSA
Mflv_4050|M.gilvum_PYR-GCK GSMAARDRMSAVSGATLSLLRDAYQRRDKVAVITFRQQDAQVLLPPTTSV
Mvan_2302|M.vanbaalenii_PYR-1 GSMAARDRMSAVSGATLSLLRDAYQRRDKVAVITFRRQDARVLLPPTTSV
MSMEG_2615|M.smegmatis_MC2_155 GSMAARDRMSAVSGAAMSLLRDAYQRRDKVAVITFRGQDARVLLPPTSSV
MAB_3157c|M.abscessus_ATCC_199 GSMAARQKMSAVSGATLSLLRDAYQRRDKVAVITFRGQDAAMLLAPTGST
TH_1386|M.thermoresistible__bu ------------------------------HVVEWSRHHRQSHLRGSRTV
*: : :. * : :.
Mb2875c|M.bovis_AF2122/97 HIAGRRLARFSTGGKTPLAEGLLAARALIIREKVRDRARRPLVVVLTDGR
Rv2850c|M.tuberculosis_H37Rv HIAGRRLARFSTGGKTPLAEGLLAARALIIREKVRDRARRPLVVVLTDGR
MMAR_1883|M.marinum_M HIASRRLARFDTGGKTPLAEGLLAARELVIREKVRDRARRPLVVVLTDGR
MUL_2111|M.ulcerans_Agy99 HIASRRLARFDTGGKTPLAEGLLAARELVIREKVRDRARRPLVVVLTDGR
MAV_3706|M.avium_104 HIAGRRLARFDTGGKTPLAEGLLAARELIVREKARDRARRALVVVLTDGR
Mflv_4050|M.gilvum_PYR-GCK HIASRRLARFDTGGKTPLAQGLLAARDMVVRERARDRARRSLVVVLTDGR
Mvan_2302|M.vanbaalenii_PYR-1 YIASRRLARFDTGGKTPLAQGLLAARDMVVRERARDRARRSLVVVLTDGR
MSMEG_2615|M.smegmatis_MC2_155 YIASRRLARFDTGGKTPLAQGLLAARDVVIREKARDRARRSLVVVLTDGR
MAB_3157c|M.abscessus_ATCC_199 HIAGRRLQRFDTGGKTPLAQGLLAARDLVARERGRDPHRRALVVVLTDGR
TH_1386|M.thermoresistible__bu GLTGSEVVALPGGGHS---------------DQQPDQHN-----------
::. .: : **:: :: * .
Mb2875c|M.bovis_AF2122/97 ATAGPDPLGRSRTAAAGLVAEGAAAVVVDCETSYVRLGLAAQLARQLGAP
Rv2850c|M.tuberculosis_H37Rv ATAGPDPLGRSRTAAAGLVAEGAAAVVVDCETSYVRLGLAAQLARQLGAP
MMAR_1883|M.marinum_M ATAGPDPLGRSRIAAARLVAEGATAVVVDCETSYVRLGLAQQLARHLGAP
MUL_2111|M.ulcerans_Agy99 ATAGPDPLGRSRIAAARLVAEGATAVVVDCETSYVRLGLAQQLARHLGAP
MAV_3706|M.avium_104 ATAGPDPLGRSRIAAARLVAEGAAAVVVDCETSYVRLGLAEHLARQLGAP
Mflv_4050|M.gilvum_PYR-GCK ATGGPDPLGRTRTAASRLVAEGAAAVVVDCETSYIRLDLAQELARQLGAP
Mvan_2302|M.vanbaalenii_PYR-1 ATGGPDPLGRARAAAARLVAEGAAAVVVDCETSYIRLDLAQDLARRLGAP
MSMEG_2615|M.smegmatis_MC2_155 ATGGPDPLQRARRAAAALVAEGTAAVVVDCETSFVRLGLAQELATQLGGE
MAB_3157c|M.abscessus_ATCC_199 ATGGRDPLGRTRRASALLRAEQVACVVIDCETSFVRMDLAVTLAQQLDAP
TH_1386|M.thermoresistible__bu ------------------QADESAHVGLRSHQDQVPTGLPMRHKSKPGRP
*: : * : .. . : .*. : .
Mb2875c|M.bovis_AF2122/97 VVRLEQLHADYLVHAVRGVA-----
Rv2850c|M.tuberculosis_H37Rv VVRLEQLHADYLVHAVRGVA-----
MMAR_1883|M.marinum_M AVRLEQLHADYLAQAIRSVA-----
MUL_2111|M.ulcerans_Agy99 AVRLEQLHADYLAQAIRSVA-----
MAV_3706|M.avium_104 AVRLEQLHADSLTRAVRDAA-----
Mflv_4050|M.gilvum_PYR-GCK AVRLEELRAEILTALVRTAA-----
Mvan_2302|M.vanbaalenii_PYR-1 SVRLDELRAENLTALVKAQADRGAA
MSMEG_2615|M.smegmatis_MC2_155 AVRLEQLRADELTRLVKTRSDAA--
MAB_3157c|M.abscessus_ATCC_199 VIQLDHLNADRLAGVVRGATAAA--
TH_1386|M.thermoresistible__bu AT------VTTLPGAVQRRPT----
. * :: .