For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PDPARARRLAKRINTIVASAIEFEIKDPGLDGVTIVDTKVTADLHDATVFYTVMGRTLDDEPDYGAAAAA LERAKGALRTMVGAGTGVRFTPTLTFTRDTTSDSVARMDELLARARAADADLARVRSGAKPAGEADPYRE SGSGVEPGRDGSIGDDDQPEY*
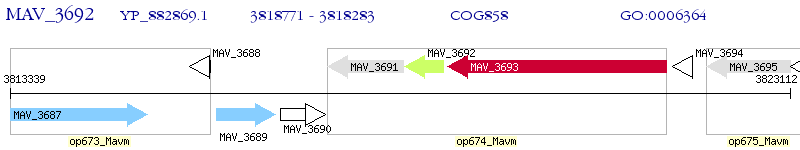
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3692 | rbfA | - | 100% (162) | ribosome-binding factor A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2863c | rbfA | 3e-64 | 77.91% (163) | ribosome-binding factor A |
| M. gilvum PYR-GCK | Mflv_4038 | rbfA | 3e-56 | 73.10% (145) | ribosome-binding factor A |
| M. tuberculosis H37Rv | Rv2838c | rbfA | 3e-64 | 77.91% (163) | ribosome-binding factor A |
| M. leprae Br4923 | MLBr_01555 | rbfA | 2e-65 | 80.62% (160) | ribosome-binding factor A |
| M. abscessus ATCC 19977 | MAB_3130c | - | 1e-53 | 72.54% (142) | ribosome-binding factor A (RbfA) |
| M. marinum M | MMAR_1895 | rbfA | 2e-68 | 84.42% (154) | ribosome-binding factor a RbfA |
| M. smegmatis MC2 155 | MSMEG_2629 | rbfA | 2e-56 | 68.55% (159) | ribosome-binding factor A |
| M. thermoresistible (build 8) | TH_2381 | rbfA | 8e-60 | 70.19% (161) | PROBABLE RIBOSOME-BINDING FACTOR A RBFA (P15B PROTEIN) |
| M. ulcerans Agy99 | MUL_2123 | rbfA | 2e-67 | 83.77% (154) | ribosome-binding factor A |
| M. vanbaalenii PYR-1 | Mvan_2315 | rbfA | 8e-56 | 70.86% (151) | ribosome-binding factor A |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_3692|M.avium_104 MPDPARARRLAKRINTIVASAIEFEIKDPGLDGVTIVDTKVTADLHDATV
MLBr_01555|M.leprae_Br4923 MADQARARRLAKRICTIVASAIEFEIKDPGLDGVTIVDVKVTADLHDATV
MMAR_1895|M.marinum_M MADPARARRLAKRITTIVASAIEYEIKDPGLVGVTITDAKVTADLHDATV
MUL_2123|M.ulcerans_Agy99 MADPARARRLAKRITTIVASAIEYEIKDPGLVGVTITDAKVTADLHDATV
Mb2863c|M.bovis_AF2122/97 MADAARARRLAKRIAAIVASAIEYEIKDPGLAGVTITDAKVTADLHDATV
Rv2838c|M.tuberculosis_H37Rv MADAARARRLAKRIAAIVASAIEYEIKDPGLAGVTITDAKVTADLHDATV
Mflv_4038|M.gilvum_PYR-GCK MPDPARAKRLAKRISTIVASAIEYEIKDPRLAGVTITDAKVTNDLHDATL
Mvan_2315|M.vanbaalenii_PYR-1 MADPARAKRLAKRISTIVASAIEYEIKDPRLAGVTITDAKVTNDLHDATL
MSMEG_2629|M.smegmatis_MC2_155 MADPARAKRLAKRISTIVASAIEYEIKDPRLAGVTITDAKVSGDLHDATL
TH_2381|M.thermoresistible__bu MADPARAKRLAKRIAAIVATAIEYEIKDPRLKGITVTDAKVTGDLRDATL
MAB_3130c|M.abscessus_ATCC_199 MADPARARRLAKRIGAIVASAIEYEIKDPRLTMVTVTDTRVTNDLHDATV
*.* ***:****** :***:***:***** * :*:.*.:*: **:***:
MAV_3692|M.avium_104 FYTVMGRTLDDEPDYGAAAAALERAKGALRTMVGAGTGVRFTPTLTFTRD
MLBr_01555|M.leprae_Br4923 FYTVMGRTLEDAPDYTAATAALNRAKGTLRSKVGAGTGVRFTPTLTFIRD
MMAR_1895|M.marinum_M YYTVMGPTLDDEPDYAAAAAALERAKGVLRTKVGAGTGVRFTPTLTFTRD
MUL_2123|M.ulcerans_Agy99 YYTVMGPTLDGEPDYAAAAAALERAKGVLRTKVGAGTGVRFTPTLTFTRD
Mb2863c|M.bovis_AF2122/97 YYTVMGRTLHDEPNCAGAAAALERAKGVLRTKVGAGTGVRFTPTLTFTLD
Rv2838c|M.tuberculosis_H37Rv YYTVMGRTLHDEPNCAGAAAALERAKGVLRTKVGAGTGVRFTPTLTFTLD
Mflv_4038|M.gilvum_PYR-GCK YYTVLGRSLDEEPDYEGAAAGLEKAKGVLRTKVGASLGVRFTPTLAFARD
Mvan_2315|M.vanbaalenii_PYR-1 YYTVLGRSLDEEPDYEGAAAGLEKAKGVLRTKVGASLGVRFTPTLAFARD
MSMEG_2629|M.smegmatis_MC2_155 YYTVLGASLDEDPDYEGAAAALEKAKGVLRTKVGAGTGVRFTPTLAFVRD
TH_2381|M.thermoresistible__bu YYTVMGRTLDEEPDYDGAAAALEKAKGVLRSKVGAGTGVRYTPTLTFVRD
MAB_3130c|M.abscessus_ATCC_199 YYTVMGQTLSDEPDFAGAAAALDKAKGVLRTKVGAGTGVRFTPTLTFVLD
:***:* :* *: .*:*.*::***.**: ***. ***:****:* *
MAV_3692|M.avium_104 TTSDSVARMDELLARARAADADLARVRSGAKPAGEADPYRESG-------
MLBr_01555|M.leprae_Br4923 TTSDSVARMEELLARARAADADVAQVRLRAKPAGEADPYRDKGSV-----
MMAR_1895|M.marinum_M VTSDTVHRMDELLARARAADADLARVRVGAKPAGEADPYRDRGSADEPSD
MUL_2123|M.ulcerans_Agy99 VTSDTVHRMDELLARARAADADLARVRVGAKPAGEADPYRDRGSVDEPSD
Mb2863c|M.bovis_AF2122/97 TISDSVHRMDELLARARAADADLARVRVGAKPAGEADPYRDNGSVAQSPA
Rv2838c|M.tuberculosis_H37Rv TISDSVHRMDELLARARAADADLARVRVGAKPAGEADPYRDNGSVAQSPA
Mflv_4038|M.gilvum_PYR-GCK TVPDAAHRMEALLAQARAADEDLARVREGAKHAGDSDPYRVLGEGDLEGP
Mvan_2315|M.vanbaalenii_PYR-1 TVPDTAHRMEALLAKARAADEDLARVRQGARHAGEADPYRVTG---AEDE
MSMEG_2629|M.smegmatis_MC2_155 TVPDAAHRMEELLARARAADEDLARVREGAKHAGDPDPYRVGG-------
TH_2381|M.thermoresistible__bu TVPDAAHRMEELLARARAADADLARVRQNAKPAGDPDPYRMDGGTGEPGA
MAB_3130c|M.abscessus_ATCC_199 TMADSARHMEELLDRTRAADAALAEVRQGAVHAGDADPYKESA-------
. .*:. :*: ** ::**** :*.** * **:.***: .
MAV_3692|M.avium_104 SGVEPGRDGS--------------IGDDDQPEY
MLBr_01555|M.leprae_Br4923 AGLVGVDIAD--------------IDDDDLTDD
MMAR_1895|M.marinum_M AGGLVIRTSDGLEAEN--------TGDDYQAED
MUL_2123|M.ulcerans_Agy99 AGGLVIRTSDGLEAEN--------TGDDYQAED
Mb2863c|M.bovis_AF2122/97 PGGLGIRTSDGPEAVEAPLTCGGDTGDDDRPKE
Rv2838c|M.tuberculosis_H37Rv PGGLGIRTSDGPEAVEAPLTCGGDTGDDDRPKE
Mflv_4038|M.gilvum_PYR-GCK ATGGPDVEDEGG------------ANSHDR---
Mvan_2315|M.vanbaalenii_PYR-1 ASG--DAEDEGG------------SNSLDD---
MSMEG_2629|M.smegmatis_MC2_155 ------AEDTDG------------DTDGDER--
TH_2381|M.thermoresistible__bu SRSGPDADDAEE------------SGDHHRRDR
MAB_3130c|M.abscessus_ATCC_199 ------AEEPAA------------YEDDERRPD
.