For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDTPSKTLLIVHHTPSPHTHEMFEAVVSGATDPEIEGVEVVRRAALTVSPAEMLAADGYLLGTPANLGYI SGALKHAFDCSYYQLLDSTRGRPFGLYLHGNEGTEGAQRAVDTITTGLGWTKAAETVVVSGKPSKDDLQA CWELGATVAAQLMD*
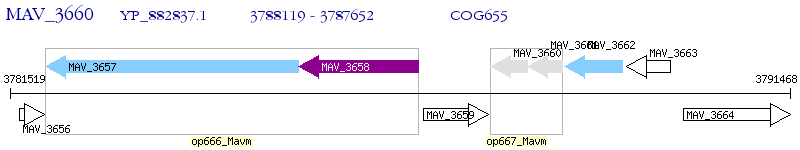
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3660 | - | - | 100% (155) | multimeric flavodoxin WrbA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2793c | - | 2e-69 | 83.78% (148) | hypothetical protein Mb2793c |
| M. gilvum PYR-GCK | Mflv_4011 | - | 8e-73 | 84.67% (150) | hypothetical protein Mflv_4011 |
| M. tuberculosis H37Rv | Rv2771c | - | 2e-69 | 83.78% (148) | hypothetical protein Rv2771c |
| M. leprae Br4923 | MLBr_01525 | - | 4e-42 | 55.33% (150) | hypothetical protein MLBr_01525 |
| M. abscessus ATCC 19977 | MAB_3094c | - | 3e-40 | 55.48% (146) | hypothetical protein MAB_3094c |
| M. marinum M | MMAR_1941 | - | 4e-68 | 82.99% (147) | hypothetical protein MMAR_1941 |
| M. smegmatis MC2 155 | MSMEG_2666 | - | 1e-44 | 57.33% (150) | multimeric flavodoxin WrbA |
| M. thermoresistible (build 8) | TH_0695 | - | 2e-69 | 80.67% (150) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2166 | - | 4e-70 | 84.56% (149) | hypothetical protein MUL_2166 |
| M. vanbaalenii PYR-1 | Mvan_2376 | - | 2e-73 | 84.67% (150) | hypothetical protein Mvan_2376 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4011|M.gilvum_PYR-GCK ----MSKTLLIVHHTPSPHCQEMFEAVLAGATDPEIEGVEVLRRPALTVS
Mvan_2376|M.vanbaalenii_PYR-1 ----MSKTLLIVHHTPSPHCHEMFEAVLAGATDPEIEGVEVLRRPALTVS
TH_0695|M.thermoresistible__bu ----MTKTLLIVHHTPSPHCQEMFEAVLSGATDPEIEGVEVLRRPALTVS
MAV_3660|M.avium_104 MTDTPSKTLLIVHHTPSPHTHEMFEAVVSGATDPEIEGVEVVRRAALTVS
Mb2793c|M.bovis_AF2122/97 -----MRRLLIVHHTPSPHMQEMFEAVVSGATDPEIEGVEVVRRPALTVS
Rv2771c|M.tuberculosis_H37Rv -----MRRLLIVHHTPSPHMQEMFEAVVSGATDPEIEGVEVVRRPALTVS
MUL_2166|M.ulcerans_Agy99 ----MSKRLLIVHHTPSPHCQEMFEAVVAGATDPEIEGVEVVRRPALTVS
MMAR_1941|M.marinum_M -----MPRLLIIHHTPSPHCQEMLEAVVAGATDPDIEGVEVVRRPALTVS
MAB_3094c|M.abscessus_ATCC_199 ----MSR-LLVVHHTPSPATRELLEAVLEGANDPEISGVEVVIRPALAAT
MSMEG_2666|M.smegmatis_MC2_155 ----MSRTLLVVHQTPSPATRELLEAVLAGARDPDITGVDVVTRPALAAT
MLBr_01525|M.leprae_Br4923 ----MTKTLLVVHHTPSPTTRELLEAVLAGANDSEIDGVEVVSRPALAAT
**::*:**** :*::***: ** *.:* **:*: *.**:.:
Mflv_4011|M.gilvum_PYR-GCK PAEMLAADGYLLGTPANLGYISGAMKHAFDCSYYQLLDATKGRPFGVFLH
Mvan_2376|M.vanbaalenii_PYR-1 PAEMLQADGYLLGTPANLGYISGALKHAFDCAYYQLLDATRGRPFGIYLH
TH_0695|M.thermoresistible__bu PVDWLAADGYLLGSPVNLGYLSGALKHAFDTAYYQILDSTSGRPFGAWLH
MAV_3660|M.avium_104 PAEMLAADGYLLGTPANLGYISGALKHAFDCSYYQLLDSTRGRPFGLYLH
Mb2793c|M.bovis_AF2122/97 PIEMLEADGYLLGTPANLGYISGALKHAFDVCYYPCLDTTRGRSFGAYIH
Rv2771c|M.tuberculosis_H37Rv PIEMLEADGYLLGTPANLGYISGALKHAFDVCYYLCLDTTRGRSFGAYIH
MUL_2166|M.ulcerans_Agy99 PVEMLEAHGYLLGTPANLGYISGALKHAFDQSYYQLLDSTRGRPFGAWMH
MMAR_1941|M.marinum_M PLEMLEADGYLLGSPVNLGYVSGALKLAFDQSYYQLLDSTRGRPFGLWLH
MAB_3094c|M.abscessus_ATCC_199 IPDMLDADGYLFGTTANFGYMSGALKHFFDTVYYPSLDHVAARPYGVWVH
MSMEG_2666|M.smegmatis_MC2_155 IPDMLDADGYLFGTTANFGYMSGALKHFFDTVYYPVLDHVAGRPYGLWVH
MLBr_01525|M.leprae_Br4923 IPDMLNADGYLFGTTANFGYMSGALKHFFDTVYYPILDHVSGRPYGLWVH
: * *.***:*:..*:**:***:* ** ** ** . .*.:* ::*
Mflv_4011|M.gilvum_PYR-GCK GNEGTEGAQRALTGITGGLGWEQAADTVVVSGKPAKNDLEACWNLGATVA
Mvan_2376|M.vanbaalenii_PYR-1 GNEGTEGAERALAGITGGLGWEQAADTVIVSGKPAKDDLEACWNLGATVA
TH_0695|M.thermoresistible__bu GNEGTEGAERALDGITGGLGWQRAAETVVVSGKPGKDDLEACWNLGATVA
MAV_3660|M.avium_104 GNEGTEGAQRAVDTITTGLGWTKAAETVVVSGKPSKDDLQACWELGATVA
Mb2793c|M.bovis_AF2122/97 GNEGTEGAERAVDAITTGLGWVQAAETVVVMGKPSKADIEACWNLGATVA
Rv2771c|M.tuberculosis_H37Rv GNEGTEGAERAVDAITTGLGWVQAAETVVVMGKPSKADIEACWNLGATVA
MUL_2166|M.ulcerans_Agy99 GNEGTEGALRAIDGITAGLGWVKAAETVVVMGKPTKADVEACWNLGATVA
MMAR_1941|M.marinum_M GNEGTEGAERAVDGITTGLGWVKAADYVVVSGKPAKADLEACWNLGATVA
MAB_3094c|M.abscessus_ATCC_199 GNNDTVGAAAAVDKIVTGLALEKAADVLEVVGPIDASTREQAYNLGGTLA
MSMEG_2666|M.smegmatis_MC2_155 GNNDTVGAANAVDRIVTGLALTKAADTLEITGAVDAAVREAAYELGGTLA
MLBr_01525|M.leprae_Br4923 GNNDTVGAAVAVGKLATGLSLTHAADVLEVVGPVDAMVCERAHELGGTLA
**:.* ** *: :. **. :**: : : * : . :**.*:*
Mflv_4011|M.gilvum_PYR-GCK ATLMD----
Mvan_2376|M.vanbaalenii_PYR-1 ATLME----
TH_0695|M.thermoresistible__bu ATLME----
MAV_3660|M.avium_104 AQLMD----
Mb2793c|M.bovis_AF2122/97 AQLMG----
Rv2771c|M.tuberculosis_H37Rv AQLMG----
MUL_2166|M.ulcerans_Agy99 AQLMSPRN-
MMAR_1941|M.marinum_M AHLMDARSR
MAB_3094c|M.abscessus_ATCC_199 AILMR----
MSMEG_2666|M.smegmatis_MC2_155 ATLMD----
MLBr_01525|M.leprae_Br4923 AMLME----
* **