For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NQPDAPTPPNDAAKALQVLKSLPSFEDTQAQVQAAMNEITAAAGKVVPSVTWETPHEGSGLGCEKPYEQT DGRGYFLPDQVAANVSVSEQQWATIQEAAKQAAAKIDATEIQVMQNNPGNHDVGFYGPTGIFIKVGYRGN LVVSGYTGCRLPRDKK*
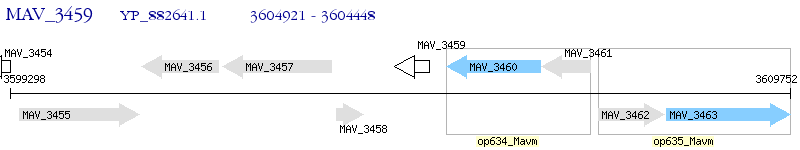
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3459 | - | - | 100% (157) | hypothetical protein MAV_3459 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2819c | lppV | 2e-14 | 27.81% (151) | lipoprotein LppV |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv2796c | lppV | 2e-14 | 27.81% (151) | lipoprotein LppV |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1914 | lppV_1 | 8e-11 | 25.83% (151) | lipoprotein LppV_1 |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3560 | - | 1e-11 | 25.00% (148) | PUTATIVE putative conserved lipoprotein LppV_1 |
| M. ulcerans Agy99 | MUL_2139 | lppV | 9e-13 | 27.81% (151) | lipoprotein LppV |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1914|M.marinum_M MRWRLVGLLALVCVLVLGCGSSGSGGLGRGGEQEGPLSPEKVAELENPLR
MUL_2139|M.ulcerans_Agy99 MRWRLVGLLALVCVLVLGCGSSGSGGLGRGGEQEGPLSPEKVAELENPLR
Mb2819c|M.bovis_AF2122/97 MRWPTAWLLALVCVMATGCGPSGHG--TRAGE-EGPLSPEKVAELENPLR
Rv2796c|M.tuberculosis_H37Rv MRWPTAWLLALVCVMATGCGPSGHG--TRAGE-EGPLSPEKVAELENPLR
TH_3560|M.thermoresistible__bu -----VLLVVLSAVVVWLSN-------ALQPPQEEPVTAEEVTALEERLR
MAV_3459|M.avium_104 -----------------------------MNQPDAPTPPNDAAKALQVLK
: * ..:..: : *:
MMAR_1914|M.marinum_M AKPSLEAAKDQYRAAVTRMADAIAALVPGLTWSMD-VDSWNHCGGEYVWT
MUL_2139|M.ulcerans_Agy99 AKPSLEAVKDQYRAAVTRMADAIAALVPGLTWSMD-VDSWNGCGGDYEWT
Mb2819c|M.bovis_AF2122/97 AKPPLEDAKDQYRAAVTQLANAITALVPGLTWRTD-MDTWTGCGGEYEWT
Rv2796c|M.tuberculosis_H37Rv AKPPLEDAKDQYRAAVTQLANAITALVPGLTWRTD-MDTWTGCGGEYEWT
TH_3560|M.thermoresistible__bu SKDSAEDTLTHYEALLAQTADQVAALVPGLTWRWNREDTTISCAGPLADT
MAV_3459|M.avium_104 SLPSFEDTQAQVQAAMNEITAAAGKVVPSVTWETPHEGSGLGCEKPYEQT
: . * . : .* : . : :**.:** .: * *
MMAR_1914|M.marinum_M N--AKMAYFMVGFSGPIPDDKWPQAVRIVKDGGQQFGATTFGVMKDKPGD
MUL_2139|M.ulcerans_Agy99 R--ARTAYFMVGFSGPIPDDKWPQAVRIVKDGGQQFGATTFGVMKDKPGD
Mb2819c|M.bovis_AF2122/97 R--AKAAYFMIVFSGPIPDDKWLQAVQIVKDGVEQFGATGFGVMKNKPAD
Rv2796c|M.tuberculosis_H37Rv R--AKAAYFMIVFSGPIPDDKWLQAVQIVKDGVEQFGATGFGVMKNKPAD
TH_3560|M.thermoresistible__bu RG-VQVLTRHALFDGPIPDEAWPHAFAIVRSQAATLGATDVFTYKDGPGD
MAV_3459|M.avium_104 DGRGYFLPDQVAANVSVSEQQWATIQEAAKQAAAKIDATEIQVMQNNPGN
. .:.:: * .:. :.** . . :: *.:
MMAR_1914|M.marinum_M HDVYLAGHGGVEFKLGTQVASSLTAQSDCRISETDTPSPPPTP-
MUL_2139|M.ulcerans_Agy99 HDVYLAGHGGVEFKLGTQVASSLTAQSDCRISETDTPSPPPTP-
Mb2819c|M.bovis_AF2122/97 HDVYFAGHGGVEFKFSTQKAAVLTAQSDCRISRTDTPKPSPTP-
Rv2796c|M.tuberculosis_H37Rv HDVYFAGHGGVEFKCSTQKAAVLTAQSDCRISRTDTPKPSPTP-
TH_3560|M.thermoresistible__bu HDVAISGDDGVEIRFGTRIAATLSARSDCRLKQADLESVSRESR
MAV_3459|M.avium_104 HDVGFYGPTGIFIKVGYRGNLVVSGYTGCRLPRDKK--------
*** : * *: :: . : ::. :.**: . .