For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGFGARLAAAKAQRGPLCVGIDPHPELLRAWDLPTTADGLAAFCDICVEAFAGFAVVKPQVAFFEAYGAA GFAVLERTIAALRSAGVLVLADAKRGDIGTTMAAYAAAWAGDSPLAADAVTASPYLGFGSLRPLLEAAAA HDRGVFVLAATSNPEGATVQRAAFDGRTVAQLVVDQAAVVNRSTNPAGPGYVGVVVGATVLQPPDLSALG GPVLVPGLGVQGGRPEALAGLGGAEPGQLLPAVAREVLRAGPDVAELRGAADRMLDAVAYLDV*
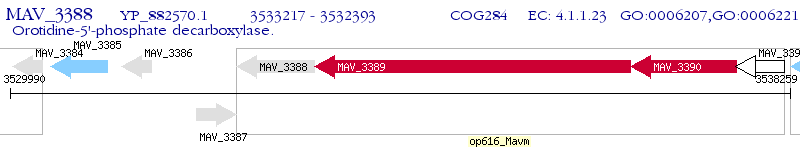
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3388 | pyrF | - | 100% (274) | orotidine 5'-phosphate decarboxylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1420 | pyrF | 1e-122 | 81.99% (272) | orotidine 5'-phosphate decarboxylase |
| M. gilvum PYR-GCK | Mflv_3742 | pyrF | 1e-117 | 77.86% (271) | orotidine 5'-phosphate decarboxylase |
| M. tuberculosis H37Rv | Rv1385 | pyrF | 1e-122 | 81.99% (272) | orotidine 5'-phosphate decarboxylase |
| M. leprae Br4923 | MLBr_00537 | pyrF | 1e-126 | 81.72% (279) | orotidine 5'-phosphate decarboxylase |
| M. abscessus ATCC 19977 | MAB_2826c | pyrF | 1e-105 | 72.86% (269) | orotidine 5'-phosphate decarboxylase |
| M. marinum M | MMAR_2200 | pyrF | 1e-126 | 82.61% (276) | orotidine 5'-phosphate decarboxylase PyrF |
| M. smegmatis MC2 155 | MSMEG_3048 | pyrF | 1e-108 | 73.29% (277) | orotidine 5'-phosphate decarboxylase |
| M. thermoresistible (build 8) | TH_0819 | pyrF | 1e-112 | 76.00% (275) | PROBABLE OROTIDINE 5'-PHOSPHATE DECARBOXYLASE PYRF (OMP |
| M. ulcerans Agy99 | MUL_1785 | pyrF | 1e-126 | 82.18% (275) | orotidine 5'-phosphate decarboxylase |
| M. vanbaalenii PYR-1 | Mvan_2665 | pyrF | 1e-116 | 78.99% (276) | orotidine 5'-phosphate decarboxylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3742|M.gilvum_PYR-GCK ---MTDSGFGERLAVARVRRGPLCLGIDPHPELLRSWNLGVDVDGLSRFA
Mvan_2665|M.vanbaalenii_PYR-1 -----MSGFGDRLAGATARRGPLCLGIDPHPELLRAWGLGTDAAGLAGFS
MAV_3388|M.avium_104 -----MTGFGARLAAAKAQRGPLCVGIDPHPELLRAWDLPTTADGLAAFC
MLBr_00537|M.leprae_Br4923 -----MTGFGARLVEATSRRGQLCLGIDPHPELLRAWDLPTTADGLAAFC
MMAR_2200|M.marinum_M -----MTGFGARLAEAKAHRGPLCLGIDPHPELLRAWDLPATADGLAAFC
MUL_1785|M.ulcerans_Agy99 -----MTGFGARLAEAKAHRGPLCLGIDPHPELLRAWDLPATADGLAAFC
Mb1420|M.bovis_AF2122/97 -----MTGFGLRLAEAKARRGPLCLGIDPHPELLRGWDLATTADGLAAFC
Rv1385|M.tuberculosis_H37Rv -----MTGFGLRLAEAKARRGPLCLGIDPHPELLRGWDLATTADGLAAFC
MSMEG_3048|M.smegmatis_MC2_155 -----MTGFGQRLDAAVSARGPLCPGIDPHPELLNAWGLTVDAEGLRAFC
TH_0819|M.thermoresistible__bu LTAAPARGFGARLADAVARRGPLCCGIDPHPELLRAWGLDTDADGLRRFS
MAB_2826c|M.abscessus_ATCC_199 ---MDPVTFGARLRAATTSRGPLCVGIDPHPELLRAWGFDADAAGLARFS
** ** * ** ** *********..*.: . . ** *.
Mflv_3742|M.gilvum_PYR-GCK DLCVTAFAGFAVVKPQVAFFEAYGSAGFAVLERTIAALRASGVLVLADAK
Mvan_2665|M.vanbaalenii_PYR-1 DICVAAFAGFAVVKPQVAFFEAYGSAGYAVLERTIAALRESGVLVLADAK
MAV_3388|M.avium_104 DICVEAFAGFAVVKPQVAFFEAYGAAGFAVLERTIAALRSAGVLVLADAK
MLBr_00537|M.leprae_Br4923 DICVEAFSGFAIVKPQVAFFEAYGAAGFAVLEYTIAALRSVGVLVLADAK
MMAR_2200|M.marinum_M DICVAAFAGFAVVKPQVAFFEAYGAAGFAVLEGTMAALRANGVLVLADAK
MUL_1785|M.ulcerans_Agy99 DICVAAFAGFAVVKPQVAFFEAYGAAGFAVLEGTMAALRANGVLVLADAK
Mb1420|M.bovis_AF2122/97 DICVRAFADFAVVKPQVAFFESYGAAGFAVLERTIAELRAADVLVLADAK
Rv1385|M.tuberculosis_H37Rv DICVRAFADFAVVKPQVAFFESYGAAGFAVLERTIAELRAADVLVLADAK
MSMEG_3048|M.smegmatis_MC2_155 DICVAAFAGFAIVKPQVAFFEAYGSAGFAVLEDTIAALRAEGVLVLADAK
TH_0819|M.thermoresistible__bu DICVTAFAELAIVKPQVAFFEAYGAAGYAVLERTIGALREAGVLVLADAK
MAB_2826c|M.abscessus_ATCC_199 DICVEAFADVAIVKPQVAFFESHGAAGYSVLERAIAGLRDAGVLVLADAK
*:** **: .*:*********::*:**::*** ::. ** .********
Mflv_3742|M.gilvum_PYR-GCK RGDIGTTMAAYATAWAGDSPLASDAVTASPYLGFGSLQPLLDTAQQNGRG
Mvan_2665|M.vanbaalenii_PYR-1 RGDIGSTMAAYASAWAGDSPLAADAVTASPYLGFGSLQPLLDTAREHDRG
MAV_3388|M.avium_104 RGDIGTTMAAYAAAWAGDSPLAADAVTASPYLGFGSLRPLLEAAAAHDRG
MLBr_00537|M.leprae_Br4923 RGDIGSTMAAYAAAWAGNSPLAADAVTASPYLGFGSLRPLLEVAAAHDRG
MMAR_2200|M.marinum_M RGDIGSTMAAYAAAWAGDSPLAADAVTVSPFLGFGSLRPLLEVATANDRG
MUL_1785|M.ulcerans_Agy99 RGDIGSTMAAYAAAWAGDSPLAADAVTVSPFLGFGSLRPLLEVATANDRG
Mb1420|M.bovis_AF2122/97 RGDIGATMSAYATAWVGDSPLAADAVTASPYLGFGSLRPLLEVAAAHGRG
Rv1385|M.tuberculosis_H37Rv RGDIGATMSAYATAWVGDSPLAADAVTASPYLGFGSLRPLLEVAAAHGRG
MSMEG_3048|M.smegmatis_MC2_155 RGDIGSTMAAYAAAWAGDSPLAADAVTASPYLGFGSLRPLLDTAVANGRG
TH_0819|M.thermoresistible__bu RGDIGSTMAAYAAAWAGDSPLAADAVTASPYLGFESLRPLLDTAAAHGRG
MAB_2826c|M.abscessus_ATCC_199 RGDIGSTMAAYAQAWLGDSPLSSDAVTASPYLGFGSLSPLLDLARERDRG
*****:**:*** ** *:***::****.**:*** ** ***: * ..**
Mflv_3742|M.gilvum_PYR-GCK VFVLAATSNPEGASVQRA----DVAGRTVAQTVVDAAAELN---------
Mvan_2665|M.vanbaalenii_PYR-1 VFVLAATSNPEGASVQCAVAPGAIPGRTVAQAIVDAAAEVN---------
MAV_3388|M.avium_104 VFVLAATSNPEGATVQRA----AFDGRTVAQLVVDQAAVVNRSTN-----
MLBr_00537|M.leprae_Br4923 VFVLASTSNLEGATVQRA----TFDGRIVAQLIVDQAAFVNREMNRSFHR
MMAR_2200|M.marinum_M VFVLAATSNPEGATIQRA----DADGRTVAQLIVDQVGLFNSEVG----E
MUL_1785|M.ulcerans_Agy99 VFVLAATSNPEGATIQRA----DADGRTVAQLIVDQVGLFNSEVG----E
Mb1420|M.bovis_AF2122/97 VFVLAATSNPEGAAVQNA----AADGRSVAQLVVDQVGAAN--------E
Rv1385|M.tuberculosis_H37Rv VFVLAATSNPEGAAVQNA----AADGRSVAQLVVDQVGAAN--------E
MSMEG_3048|M.smegmatis_MC2_155 VFVLAATSNPEGVGLQRA----VAGDVTVAQSIVDAVAQANREADP--AA
TH_0819|M.thermoresistible__bu VFVLAATSNPEGVPLQRA----RVGDRTVAQWIVDSAAAENQAAS--AAG
MAB_2826c|M.abscessus_ATCC_199 VFVLAATSNPEAPPVQRA----RAGEKTVAQAMVDQVAALN---------
*****:*** *. :* * *** :** .. *
Mflv_3742|M.gilvum_PYR-GCK -SPGAPGSVGVVVGATLSDPPDVSALNGPVLVPGVGAQGGRPEALAGLAG
Mvan_2665|M.vanbaalenii_PYR-1 -AGAARGSVGVVVGATLSAPPDVSALNGPVLVPGVGAQGGRPEALAGLGG
MAV_3388|M.avium_104 --PAGPGYVGVVVGATVLQPPDLSALGGPVLVPGLGVQGGRPEALAGLGG
MLBr_00537|M.leprae_Br4923 SEPGCLGYVGVVVGATVFGAPDVSALGGPVLVPGVGAQGGHPEALGGLGG
MMAR_2200|M.marinum_M VTGPEPGSVGVVVGATVLNPPDVSGLGGPVLVPGVGVQGGRPEALGGLGG
MUL_1785|M.ulcerans_Agy99 VTGSEPGSVGVVVGATVLNPPDVSGLGGPVLVPGVGVQGGRPEALGGLGG
Mb1420|M.bovis_AF2122/97 AAGPGPGSIGVVVGATAPQAPDLSAFTGPVLVPGVGVQGGRPEALGGLGG
Rv1385|M.tuberculosis_H37Rv AAGPGPGSIGVVVGATAPQAPDLSAFTGPVLVPGVGVQGGRPEALGGLGG
MSMEG_3048|M.smegmatis_MC2_155 RDGDPVGPFGVVVGATVADPPDLHMLGGPVLVPGVGAQGGRPEALGGLGN
TH_0819|M.thermoresistible__bu ANQAASGSVGVVVGATVAEPPDLSGLGGPVLVPGVGAQGGRPEQLSGLGG
MAB_2826c|M.abscessus_ATCC_199 AEQAGPGSVGVVVGVTVTDPPDLSSLGGPVLVPGLGAQGGKPEDLRGLGG
* .*****.* .**: : *******:*.***:** * **..
Mflv_3742|M.gilvum_PYR-GCK AGPGQLLPAVSREVLRSGPDVAALRDAAERMRDAVAYLA---
Mvan_2665|M.vanbaalenii_PYR-1 ARRGQLLPAVSREVLRAGPDVAALRAAAERMRDAVAYLGENP
MAV_3388|M.avium_104 AEPGQLLPAVAREVLRAGPDVAELRGAADRMLDAVAYLDV--
MLBr_00537|M.leprae_Br4923 AAPGQLLPAVSRAVLRAGPGVSELRAAGEQMRDAVAYLAAV-
MMAR_2200|M.marinum_M AAPQQLLPAVSREVLRAGPSISEVRAAAERMLDSVAYLSAV-
MUL_1785|M.ulcerans_Agy99 AAPQQLLPAVSREVLRAGPSISEVRAAAERMLDSVAYLSAV-
Mb1420|M.bovis_AF2122/97 AASSQLLPAVAREVLRAGPGVPELRAAGERMRDAVAYLAAV-
Rv1385|M.tuberculosis_H37Rv AASSQLLPAVAREVLRAGPGVPELRAAGERMRDAVAYLAAV-
MSMEG_3048|M.smegmatis_MC2_155 AR--RLLPAVSREVLRAGPAVDDVRAAAERLRDQVAYLA---
TH_0819|M.thermoresistible__bu AGRGQLLPAVSRAVLRSGPDPTALRSAAERIRDAVAHLA---
MAB_2826c|M.abscessus_ATCC_199 AP-GSLLPAVSREVLRAGPDATALRAQVSRLRDSVAYLLD--
* *****:* ***:** :* .:: * **:*