For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTAQTGRRVAISAGSLAVLLGALDAYVVVTIMRDIMTDVHIPINQLQRITWIITMYLLGYIAAMPLLGRA SDRFGRKLVLQVSLALFMVGSVVTALAGHWGDFHLLIGGRTIQGVASGALLPVTLALGADLWAQRNRAGV LGGIGAAQELGSVLGPLYGIFIVFLFHDWRYVFWINVPLTLIAMVMIQFSLPSHEKVEQPEKVDLVGGVL LAVALGLAVIGLYNPEPDGKQILPSYGLPLVLGAVVVGILFLLWERFARTRLIEPAGVHFRPFLAALGAS LFAGAALMVTLVDVELFGQGVLGQDQTQAAGLLLWFLIALPIGAVLGGWIATRVGDRAMTFVGLLIAAYG YWLIHYWRQDVLSQKHNVLGLFSVPVLHADLLVAGVGLGLVIGPLTSAALRVVPSAQHGIASAAVVVARM TGMLIGVAALSAWGLYRFNQIVANLTAAIPPNASLLERIAAQGTMYLKAFAMMYGDIFAATVVICIAGAL LGLLIGGRKEHAEEPEIVEPQAVSLGER
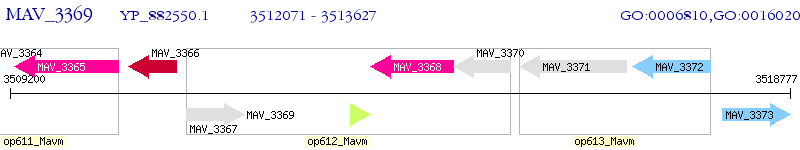
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3369 | - | - | 100% (518) | aminoglycosides/tetracycline-transport integral membrane protein |
| M. avium 104 | MAV_1387 | - | 6e-32 | 28.13% (423) | drug transporter |
| M. avium 104 | MAV_3702 | - | 1e-30 | 28.67% (422) | transporter, major facilitator family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1445c | - | 0.0 | 80.83% (506) | aminoglycosides/tetracycline-transport integral membrane |
| M. gilvum PYR-GCK | Mflv_3725 | - | 0.0 | 68.24% (507) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv1410c | - | 0.0 | 80.83% (506) | aminoglycosides/tetracycline-transport integral membrane |
| M. leprae Br4923 | MLBr_00556 | - | 0.0 | 76.23% (509) | putative integral membrane drug transport protein |
| M. abscessus ATCC 19977 | MAB_2807 | - | 1e-166 | 60.04% (503) | major facilitator transporter |
| M. marinum M | MMAR_2219 | - | 0.0 | 79.18% (514) | aminoglycosides/tetracycline-transport integral membrane |
| M. smegmatis MC2 155 | MSMEG_3069 | - | 0.0 | 68.24% (510) | aminoglycosides/tetracycline-transport integral membrane |
| M. thermoresistible (build 8) | TH_1008 | - | 0.0 | 67.27% (498) | AMINOGLYCOSIDES/TETRACYCLINE-TRANSPORT INTEGRAL MEMBRANE |
| M. ulcerans Agy99 | MUL_1806 | - | 0.0 | 78.79% (514) | aminoglycosides/tetracycline-transport integral membrane |
| M. vanbaalenii PYR-1 | Mvan_2691 | - | 0.0 | 69.41% (510) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1445c|M.bovis_AF2122/97 --MRAG--------------RRVAISAGSLAVLLGALDTYVVVTIMRDIM
Rv1410c|M.tuberculosis_H37Rv --MRAG--------------RRVAISAGSLAVLLGALDTYVVVTIMRDIM
MMAR_2219|M.marinum_M MSTQTG--------------RRVAISAGSLAVLLGALDTYVVVTIMRDIM
MUL_1806|M.ulcerans_Agy99 MSTQTG--------------RRVAISAGSLAVLLGALDTYVVVTIMRDIM
MLBr_00556|M.leprae_Br4923 MSTRAG--------------RRVAISAGSLAVLLGALDTYVVVTIMRDIM
MAV_3369|M.avium_104 MTAQTG--------------RRVAISAGSLAVLLGALDAYVVVTIMRDIM
Mflv_3725|M.gilvum_PYR-GCK ---MPTITAPA------ATNRLIAISAGSLAVILGALDTYVVVTIMVDIM
Mvan_2691|M.vanbaalenii_PYR-1 MHRTPITAKPAPHAGRGAINRRIAISAGSLAVVLGALDTYVVVTIMVDIM
TH_1008|M.thermoresistible__bu MRRATAAAIRPPR----GHNRTIAISAGSLAVLLGALDTYVVITIMVDIM
MSMEG_3069|M.smegmatis_MC2_155 MSSRG--------------NRNIAISAGSLAVLLGALDTYVVITIIVDIM
MAB_2807|M.abscessus_ATCC_1997 MTHTATEPGTSLP----LRHRSIAIGAGSLAVLLGALDTYVVVSVITDIM
* :**.******:*****:***:::: ***
Mb1445c|M.bovis_AF2122/97 NSVGIPINQLHRITWIVTMYLLGYIAAMPLLGRASDRFGRKLMLQVSLAG
Rv1410c|M.tuberculosis_H37Rv NSVGIPINQLHRITWIVTMYLLGYIAAMPLLGRASDRFGRKLMLQVSLAG
MMAR_2219|M.marinum_M GSVGIPVNQLQRITWIVTWYLLGYIAAMPLLGRASDRFGRKLMLQVSLLG
MUL_1806|M.ulcerans_Agy99 GSVGIPVNQLQRITWIVTWYLLGYIAAMPLLGRASDRFGRKLMLQVSLLG
MLBr_00556|M.leprae_Br4923 HDVGIPVNQMQRITWIVTMYLLGYIAAMPLLSRASDRFGRKLLLQVSLAG
MAV_3369|M.avium_104 TDVHIPINQLQRITWIITMYLLGYIAAMPLLGRASDRFGRKLVLQVSLAL
Mflv_3725|M.gilvum_PYR-GCK SDVGIGINEIQRVTPIITGYLLGYIAAMPLLGRASDRFGRKMLIQVSLAG
Mvan_2691|M.vanbaalenii_PYR-1 RDVGIGVNEIQRVTPIITGYLLGYIAAMPLLGRASDRFGRKILIQVSLAG
TH_1008|M.thermoresistible__bu RDVGIALNKIHQVTPIITGYLLGYIAAMPLLGRASDRFGRKMLIQVGLAG
MSMEG_3069|M.smegmatis_MC2_155 ADVGIAINQIQQVTPIITGYLLGYIAAMPLLGRASDRFGRKMLIQVGLAG
MAB_2807|M.abscessus_ATCC_1997 TSVGIALNNIQQVTPIVTGYLLGYIAAMPLLGQASDRFGRRLILQLALAG
.* * :*:::::* *:* ************.:*******::::*:.*
Mb1445c|M.bovis_AF2122/97 FIIGSVVTALAGHFGDFHMLIAGRTIQGVASGALLPITLALGADLWSQRN
Rv1410c|M.tuberculosis_H37Rv FIIGSVVTALAGHFGDFHMLIAGRTIQGVASGALLPITLALGADLWSQRN
MMAR_2219|M.marinum_M FAAGSVVTALAGHFGDFHMLLAGRALQGVASGALLPITLALGADLWAQRN
MUL_1806|M.ulcerans_Agy99 FAAGSVVTALAGHFGDFHMLLAGRALQGVASGALLPITLALGADLWAQRN
MLBr_00556|M.leprae_Br4923 FAIGSVMTALAGQFGDFHMLIAGRTIQGVASGALLPITLALGADLWAQRN
MAV_3369|M.avium_104 FMVGSVVTALAGHWGDFHLLIGGRTIQGVASGALLPVTLALGADLWAQRN
Mflv_3725|M.gilvum_PYR-GCK FALGSVVTAMSTDLD---ILVLGRVIQGTASGALLPVTLALAADLWAARN
Mvan_2691|M.vanbaalenii_PYR-1 FAAGSVITAMSTDLV---VLVVGRVIQGTASGALLPVTLALAADLWAARN
TH_1008|M.thermoresistible__bu FAIGSVVTALAGDLQSLHMLVTGRIIQGAASGALLPVTLALAADLWAERN
MSMEG_3069|M.smegmatis_MC2_155 FAVGSVVTALSSDLT---MLVIGRIIQGSASGALLPVTLALAADLWSARS
MAB_2807|M.abscessus_ATCC_1997 FAIGSVITALSTDLT---MLVSGRVIQGVASGALLPVTLALGADLWSQRN
* ***:**:: . :*: ** :** *******:****.****: *.
Mb1445c|M.bovis_AF2122/97 RAGVLGGIGAAQELGSVLGPLYGIFIVWLLHDWRDVFWINVPLTAIAMVM
Rv1410c|M.tuberculosis_H37Rv RAGVLGGIGAAQELGSVLGPLYGIFIVWLLHDWRDVFWINVPLTAIAMVM
MMAR_2219|M.marinum_M RAGVLGGIGAAQELGSVLGPLYGIFIVWLFHDWRDVFWINVPLTAIAMLM
MUL_1806|M.ulcerans_Agy99 RAGVLGGIGAAQELGSVLGPLYGIFIVWLFHDWRDVFWINVPLTAIAMLM
MLBr_00556|M.leprae_Br4923 RAGVLGGIGAAQELGSVLGPLYGIFIVWLFSDWRYVFWINIPLTAIAMLM
MAV_3369|M.avium_104 RAGVLGGIGAAQELGSVLGPLYGIFIVFLFHDWRYVFWINVPLTLIAMVM
Mflv_3725|M.gilvum_PYR-GCK RAAVLGWVGAAQELGSVLGPIYGISLVWLFHHWQSVFWVNVPLALVAMVM
Mvan_2691|M.vanbaalenii_PYR-1 RAAVLGGVGAAQELGSVLGPVYGISLVWLFGHWQSVFWVNVPLAVIAMVM
TH_1008|M.thermoresistible__bu RAGVLGGIGAAQELGAVLGPLYGIAMVWLFGAWESVFWVNVPLAALAMVM
MSMEG_3069|M.smegmatis_MC2_155 RASVLGGVGAAQELGAVLGPMYGIALVWLFNHWQAVFWVNVPLAVIAMVM
MAB_2807|M.abscessus_ATCC_1997 RATVLGGIGAAQELGSVLGPLYGVLCVWLFGSWTAIFWVNVPLAIIAIVL
** *** :*******:****:**: *:*: * :**:*:**: :*:::
Mb1445c|M.bovis_AF2122/97 IHFSLPSHDRSTEPERVDLVGGLLLALALGLAVIGLYNPNPDGKHVLPDY
Rv1410c|M.tuberculosis_H37Rv IHFSLPSHDRSTEPERVDLVGGLLLALALGLAVIGLYNPNPDGKHVLPDY
MMAR_2219|M.marinum_M IQFSLPAHDKNAEPEKIDLVGGALLAIALGLAVIGLYNPNPDGKEVLPSY
MUL_1806|M.ulcerans_Agy99 IQFSLPAHDKNAEPEKIDLVGGALLAIALGLAVIGLYNPNPDGKEVLPSY
MLBr_00556|M.leprae_Br4923 IQVSLSAHDRGDELEKVDVVGGVLLAIALGLVVIGLYNPQPDSKQVLPSY
MAV_3369|M.avium_104 IQFSLPSHEKVEQPEKVDLVGGVLLAVALGLAVIGLYNPEPDGKQILPSY
Mflv_3725|M.gilvum_PYR-GCK IHFSLPGRMETEVPQKIDVVGGLLLAVALGLAVIGLYNPKPDGKEVLPSW
Mvan_2691|M.vanbaalenii_PYR-1 IHFSLPGRSPTEDPQKVDVVGGLLLAVALGLAVIGLYNPKPDGKTVLPSW
TH_1008|M.thermoresistible__bu IHFSLPARDPDGPRERIDVVGGVLLGITLFLIVVGLYNPQPSAREVVPTW
MSMEG_3069|M.smegmatis_MC2_155 IHFSLPARQQVDEPERVDVIGGVLLAIALGLTVVGLYNPEPDGKQVLPSW
MAB_2807|M.abscessus_ATCC_1997 VQFSVPAHQHDPDRPKVDVVGGALLAVALGLLVVGLYSPDPK-VSALPEW
::.*:..: ::*::** **.::* * *:***.*.*. :* :
Mb1445c|M.bovis_AF2122/97 GAPLLVGALVAAVAFFGWERFARTRLIDPAGVHFRPFLSALGASVAAGAA
Rv1410c|M.tuberculosis_H37Rv GAPLLVGALVAAVAFFGWERFARTRLIDPAGVHFRPFLSALGASVAAGAA
MMAR_2219|M.marinum_M GLPVLTGAIVAAVAFFAWERFARTRLIDPNGVHFRPFLAALGSSVAAGAA
MUL_1806|M.ulcerans_Agy99 GLPVLTGAIVAAVAFFAWERFARTRLIDPNGVHFRPFLAALGSSVAAGAA
MLBr_00556|M.leprae_Br4923 GVPVLVGGIVATVAFAVWERCARTRLIDPAGVHIRPFLSALGASVAAGAA
MAV_3369|M.avium_104 GLPLVLGAVVVGILFLLWERFARTRLIEPAGVHFRPFLAALGASLFAGAA
Mflv_3725|M.gilvum_PYR-GCK GVPVLIAALIAAVAFFVWEKFARTRLLDPKGVRFRPFLAALGASLAAGAA
Mvan_2691|M.vanbaalenii_PYR-1 GLPVLIAAAVAAVAFFGWEKFARTRLLEPRGVHFRPFLAALGASLTAGAA
TH_1008|M.thermoresistible__bu RLWLLAAAAVTLIGFFVWERRARTKLIDPAGVRFRPFLAALGASFVAGAA
MSMEG_3069|M.smegmatis_MC2_155 GLPVLAGALVAAVAFFAWEKVAKTRLIDPAGVRFRPFLAALAASLCAGAA
MAB_2807|M.abscessus_ATCC_1997 ALPVLSGSGVAFLAFILWESRAKTRLIKPEGVRFGPFFAALAASLAAGAA
:: .. :. : * ** *:*:*:.* **:: **::**.:*. ****
Mb1445c|M.bovis_AF2122/97 LMVTLVDVELFGQGVLQMDQAQAAGMLLWFLIALPIGAVTGGWIATRAGD
Rv1410c|M.tuberculosis_H37Rv LMVTLVDVELFGQGVLQMDQAQAAGMLLWFLIALPIGAVTGGWIATRAGD
MMAR_2219|M.marinum_M LMVTLVNVELFGQGVLSMDQTQAAGMLLWFLIALPIGAVTGGWIATRVGD
MUL_1806|M.ulcerans_Agy99 LMVTLVNVELFGQGVLSMDQTQAARMLLWFLIALPIGAVTGGWIATRVGD
MLBr_00556|M.leprae_Br4923 LMVTLVNVELFGQGVLGMDQTQAAGLLVWFLIALPIGAVLGGWGATKAGD
MAV_3369|M.avium_104 LMVTLVDVELFGQGVLGQDQTQAAGLLLWFLIALPIGAVLGGWIATRVGD
Mflv_3725|M.gilvum_PYR-GCK LMVTLVNVELFAQGVLGKDQIEAAFLLLWFLAALPVGALLGGWLATRVGD
Mvan_2691|M.vanbaalenii_PYR-1 LMVTLVNVELFGQGVLGQDQNEAAFLLLRFLIALPVGALLGGWLATRVGD
TH_1008|M.thermoresistible__bu LMVTLVNVELFGQGVLGKDQFEAAMLLLRFLIALPIGALIGGWLATRVGD
MSMEG_3069|M.smegmatis_MC2_155 LMVTLVNVELFGQGVLGQDQDHAAFLLLRFLIALPIGALIGGWLATRIGD
MAB_2807|M.abscessus_ATCC_1997 LMVTLVNIELLGQGVLQMDKADAVFLLSRFLVALPIGAVIGGWLATRFGD
******::**:.**** *: .*. :* ** ***:**: *** **: **
Mb1445c|M.bovis_AF2122/97 RAVAFAGLLIAAYGYWLISHWPVDLLADRHNILGLFTVPAMHTDLVVAGL
Rv1410c|M.tuberculosis_H37Rv RAVAFAGLLIAAYGYWLISHWPVDLLADRHNILGLFTVPAMHTDLVVAGL
MMAR_2219|M.marinum_M RAVTFVGLLIAAGGYLLISHWPVDLPDYRHSIFGLFTVPAMHADLLVAGL
MUL_1806|M.ulcerans_Agy99 RAVTFVGLLIAAGGYLLISHWPVDLPDYRHSIFGLFTVPAMHADLLVAGL
MLBr_00556|M.leprae_Br4923 RTMTFVGLLITAGGYWLISHWPVDLLNYRRSIFGLFSVPTMYADLLVAGL
MAV_3369|M.avium_104 RAMTFVGLLIAAYGYWLIHYWRQDVLSQKHNVLGLFSVPVLHADLLVAGV
Mflv_3725|M.gilvum_PYR-GCK RIIAFAGLLIAAGGYLLISHWPADVLEARHS-IGFLSLPVLGTDLIITGF
Mvan_2691|M.vanbaalenii_PYR-1 RLVTFAGLLIAAGGFLLISKWPVDLLAARHD-LGFVSLPVLDTDLAIAGL
TH_1008|M.thermoresistible__bu RIVTFVGLLIAAGGFWLISHWPVDLLAARHD-LGVVTLPVLDTDLAIVGI
MSMEG_3069|M.smegmatis_MC2_155 RLVVLIGLLIAAGGFVLISHWSVDVLADRHN-LGLFTLPVLDTDLAIVGL
MAB_2807|M.abscessus_ATCC_1997 RIIAVIGMLIAAFGYYLISGWPVDVLAAVHN-FGFFTLPRLDTDLVVAGI
* :.. *:**:* *: ** * *: :. :*..::* : :** :.*.
Mb1445c|M.bovis_AF2122/97 GLGLVIGPLSSATLRVVPSAQHGIASAAVVVARMTGMLIGVAALSAWGLY
Rv1410c|M.tuberculosis_H37Rv GLGLVIGPLSSATLRVVPSAQHGIASAAVVVARMTGMLIGVAALSAWGLY
MMAR_2219|M.marinum_M GLGLVIGPLSSATLRVVPAAQHGIASAAVVVARMTGMLIGVAALSAWGLY
MUL_1806|M.ulcerans_Agy99 GLGLVIGPLSSATLRVLPAAQHGIASAAVVVARMTGMLIGVAALSAWGLY
MLBr_00556|M.leprae_Br4923 GLGLVIGPLSSATLRVVPTAQHGIASAAVVVARMTGMLIGVAALTAWGLY
MAV_3369|M.avium_104 GLGLVIGPLTSAALRVVPSAQHGIASAAVVVARMTGMLIGVAALSAWGLY
Mflv_3725|M.gilvum_PYR-GCK GLGLVIGPLTSATLRVVPSAQHGIASAAVVVSRMIGMLIGIAALSAWGLY
Mvan_2691|M.vanbaalenii_PYR-1 GLGLVIGPLTSATLRVVPAAQHGIASAAVVVARMIGMLIGIAALSAWGLY
TH_1008|M.thermoresistible__bu GLGLVIGPLTSAALRVVPAAEHGIASAAVVVARMIGMVIGLAALSAWGLY
MSMEG_3069|M.smegmatis_MC2_155 GLGLVIGPLTSATLRAVPAAEHGIASAAVVVARMIGMLIGIAALGAWGFY
MAB_2807|M.abscessus_ATCC_1997 GLGLVIGPLSSAALRVVPAAQHGIASALVVVARMTGMLIGMAALGGWGIH
*********:**:**.:*:*:****** ***:** **:**:*** .**::
Mb1445c|M.bovis_AF2122/97 RFNQILAGLSAAIPPNAS-----LLERAAAIGARYQQAFALMYGEIFTIT
Rv1410c|M.tuberculosis_H37Rv RFNQILAGLSAAIPPNAS-----LLERAAAIGARYQQAFALMYGEIFTIT
MMAR_2219|M.marinum_M RFNQILAGLSAAVPPDAT-----LLERAAAIGAQYQQAFALMYGEIFTIT
MUL_1806|M.ulcerans_Agy99 RFNQILAGLSAAVPPDAT-----LLERAAAIGAQYQQAFALMYGEIFTIT
MLBr_00556|M.leprae_Br4923 RFNQILAGLSTAVPPDAT-----LIERAAAVADQAKRAYTMMYSDIFMIT
MAV_3369|M.avium_104 RFNQIVANLTAAIPPNAS-----LLERIAAQGTMYLKAFAMMYGDIFAAT
Mflv_3725|M.gilvum_PYR-GCK RFNQHLASLPAS-AGGDS-----LAERLAAEANRVREAYVLQYAEIFTIT
Mvan_2691|M.vanbaalenii_PYR-1 RFNQHLASLPAS-TAGGS-----LAERLAAEANRVREAYVLQYGEIFTIT
TH_1008|M.thermoresistible__bu RFNQHLQSLAAREPGGDS-----LAARLAAEAARYREAYAMQYGEIFAIT
MSMEG_3069|M.smegmatis_MC2_155 RFNQHLATLAAR-AAGDAGSPMSLAERLTAQAVRYREAYVMMYGDIFLSA
MAB_2807|M.abscessus_ATCC_1997 RFYQNFDALAAKEPKPTGKP--NLLELQQQLLNRSITAYSEMYSEMFAIT
** * . *.: . * *: *.::* :
Mb1445c|M.bovis_AF2122/97 AIVCVFGAVLGLLISGRKEHADEP--------------------------
Rv1410c|M.tuberculosis_H37Rv AIVCVFGAVLGLLISGRKEHADEP--------------------------
MMAR_2219|M.marinum_M AFVCVFGAVLGLLVSGRKEHAEEP--------------------------
MUL_1806|M.ulcerans_Agy99 AFVCVFGAVLGLLVSGRKEHAEEP--------------------------
MLBr_00556|M.leprae_Br4923 AIVCVIGALLGLLISSRKEHASEP--------------------------
MAV_3369|M.avium_104 VVICIAGALLGLLIGGRKEHAEEP--------------------------
Mflv_3725|M.gilvum_PYR-GCK AVVCVVGAVLGLLIAARDTHAEET--------------------------
Mvan_2691|M.vanbaalenii_PYR-1 AVVCLVGALLGLLIAAKDDHAEE---------------------------
TH_1008|M.thermoresistible__bu ALLCVAGALLGLLIAGHGDRADDTGTPDGSVWDLHGSGDDDTPTRVIGRQ
MSMEG_3069|M.smegmatis_MC2_155 AVVCVIGALLGLLISGKHEHAEEFEPAYAPTYGGGGAIDPYDAGDADDAP
MAB_2807|M.abscessus_ATCC_1997 AVVCVIAAVIAIFVGSHRKSGHEAQ-------------------------
..:*: .*::.:::..: . :
Mb1445c|M.bovis_AF2122/97 -EVQEQPTLAPQVEPL-------------
Rv1410c|M.tuberculosis_H37Rv -EVQEQPTLAPQVEPL-------------
MMAR_2219|M.marinum_M -EISEEPAVAPRP----------------
MUL_1806|M.ulcerans_Agy99 -EISEEPAVAPRP----------------
MLBr_00556|M.leprae_Br4923 -KVPE------------------------
MAV_3369|M.avium_104 -EIVEPQAVSLGER---------------
Mflv_3725|M.gilvum_PYR-GCK DPVENAQQRS-SPRPIS------------
Mvan_2691|M.vanbaalenii_PYR-1 -PSDEAQQRS-SPGPIS------------
TH_1008|M.thermoresistible__bu VPLPSMEQTVRMPRPPGD-----------
MSMEG_3069|M.smegmatis_MC2_155 TEMLDLPTQVLSAPPSDPGDERPGRHRAP
MAB_2807|M.abscessus_ATCC_1997 -----------------------------