For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LGHYIANVRDLEFNLFEVLDVGAVLGSGGYGDLDEETARTILAEAARLAEGPVAETFAFADRNPPVFDPA EHAITVPEELAKTVAALKEAGWWRLGMAEEIGGMPAPPPLAWAVNEMLICANPSANFFCLGPFMSQALYL EGNEQQRRWAAEGVERGWAATMVLTEPDAGSDVGAGRAKAIEQPDGTWHIEGVKRFISGGDVGDTAENIF HLVLARPEGAGPGTKGLSLFYVPKYLFDPETFELGPRNGVFVTGVEHKMGIKSSPTCELTFGATDVPAVG YLVGDVHRGIAQMFTVIEQARMTIGVKAAGTLSTGYLNALAFAKERVQGADLTQMADKTAPRVTIIHHPD VRRSLMTQKAYAEGLRALYMYAAAHQDDQVAQRVSGADHDMAHRVDDLLLPIVKGVSSERAYEVLTESLQ TLGGSGFLQDYPLEQYIRDSKIDSLYEGTTAIQALDFFFRKIVRDRGQALQFVTAQITATVDDCDEALRP IAELLQTAFDDVTAMTGALTGYLMSAAEQPTDIYKVGLASVRYLLAVGDLLIGWRLLVHADVAQRALAAG AGGDDEAFYRGKIATAAFFAKNMLPRLSGLRGIIEAIDDEIMKLPEAAF
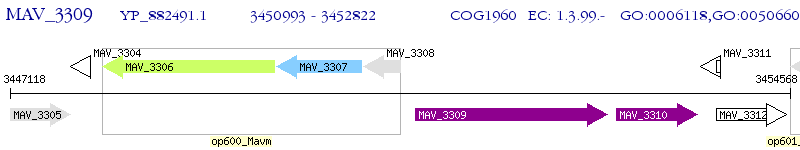
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3309 | - | - | 100% (609) | putative acyl-CoA dehydrogenase |
| M. avium 104 | MAV_4914 | - | 0.0 | 64.16% (611) | putative acyl-CoA dehydrogenase |
| M. avium 104 | MAV_4243 | - | 2e-28 | 28.53% (389) | acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1502c | fadE15 | 0.0 | 84.24% (609) | acyl-CoA dehydrogenase FADE15 |
| M. gilvum PYR-GCK | Mflv_0430 | - | 0.0 | 64.44% (613) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv1467c | fadE15 | 0.0 | 84.24% (609) | acyl-CoA dehydrogenase FADE15 |
| M. leprae Br4923 | MLBr_02563 | fadE5 | 0.0 | 63.01% (611) | acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4437 | - | 0.0 | 62.19% (611) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_2273 | fadE15 | 0.0 | 84.40% (609) | acyl-CoA dehydrogenase FadE15 |
| M. smegmatis MC2 155 | MSMEG_0406 | - | 0.0 | 65.47% (611) | acyl-CoA-dehydrogenase |
| M. thermoresistible (build 8) | TH_1915 | fadE15 | 0.0 | 65.85% (612) | PROBABLE ACYL-CoA DEHYDROGENASE FADE15 |
| M. ulcerans Agy99 | MUL_1869 | fadE15 | 0.0 | 84.40% (609) | acyl-CoA dehydrogenase FadE15 |
| M. vanbaalenii PYR-1 | Mvan_0238 | - | 0.0 | 64.48% (611) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4437|M.abscessus_ATCC_1997 MTHMGHYKSNVRDLEFNLFELFKIQQVFGGEEFPELDEDTARTFLAEMRT
MSMEG_0406|M.smegmatis_MC2_155 ---MSHYKSNVRDQVFNLFEVFGVDKVLGADKFSDLDADTAREMLTEIAR
MLBr_02563|M.leprae_Br4923 ---MSHYKSNVRDQVFNLFEVLGVDKAFGTDEFSNLDADTAYEMLTEVSR
TH_1915|M.thermoresistible__bu ---VSHYKSNVRDQEFNLFEVFGVDKALGQGEYSDLDVDTAREMLAEISR
Mflv_0430|M.gilvum_PYR-GCK ---MGHYKSNVRDLEFNLFEVLALEKALATGDFGDLDLQSARDMLAQAVE
Mvan_0238|M.vanbaalenii_PYR-1 ---MGHYKSNVRDLEFNLFEVLALEKVLATGEFGDLDGPSVREMLHEAAK
Mb1502c|M.bovis_AF2122/97 ---MGHYIANVRDLEFNLLEVLDIGAVLGTGRYSDLDVDTVRTILAEAAR
Rv1467c|M.tuberculosis_H37Rv ---MGHYIANVRDLEFNLLEVLDIGAVLGTGRYSDLDVDTVRTILAEAAR
MMAR_2273|M.marinum_M ---MGHYIANVRDLEFNLFEVLDVGAVLGTGKYSELDADTVRTILSEAAR
MUL_1869|M.ulcerans_Agy99 ---MGHYIANVRDLEFNLFEVLDVGAVLGTGKYSELDADTVRTILSEAAR
MAV_3309|M.avium_104 ---MGHYIANVRDLEFNLFEVLDVGAVLGSGGYGDLDEETARTILAEAAR
:.** :**** ***:*:: : .:. : :** :. :* :
MAB_4437|M.abscessus_ATCC_1997 LAEGPLADSFAEGDRNPPVFDPETHSVAIPEAFKKSVKAITEAGWDRVGL
MSMEG_0406|M.smegmatis_MC2_155 LAEGPIAESFVEGDRNPPVFDPETHTVTLPEGFKKSMRALFDGGWDKVGL
MLBr_02563|M.leprae_Br4923 LAEGPVAASFAEGDRNPPVFHPETHSITLPESFKKSCRAVAEAGWSKAGI
TH_1915|M.thermoresistible__bu LAEGPVAASFVEGDRNPPTFDPETHSVSLPENFKKSVRAVLDAGWDKVGI
Mflv_0430|M.gilvum_PYR-GCK LAEGPVAEAFSETDRRPPSFDPATHVVSIPEPFKKSFRAWYDGEWFRVGM
Mvan_0238|M.vanbaalenii_PYR-1 LAEGPLAEAYAETDRNPPSFDPATHVVSIPEPFKKSFRAWLDGEWFRVGM
Mb1502c|M.bovis_AF2122/97 LAEGPIAESFGYADRNPPVFDPNTHSISVPDELAKTVQAIKEAGWWRLGL
Rv1467c|M.tuberculosis_H37Rv LAEGPIAESFGYADRNPPVFDPNTHSISVPDELAKTVQAIKEAGWWRLGL
MMAR_2273|M.marinum_M LAEGPVAETFAAADRNPPVFDPATHSISVPDELVKTVQAIKDAEWWRLGL
MUL_1869|M.ulcerans_Agy99 LAEGPVAETFAAADRNPPVFDPATHSISVPDELVKTVQAIKDAEWWRLGL
MAV_3309|M.avium_104 LAEGPVAETFAFADRNPPVFDPAEHAITVPEELAKTVAALKEAGWWRLGM
*****:* :: **.** *.* * :::*: : *: * :. * : *:
MAB_4437|M.abscessus_ATCC_1997 QEELGGTPVPRSLSWAIQEMILGANPAVWMYSGGAGFAQIFYNIATDEQK
MSMEG_0406|M.smegmatis_MC2_155 AEHLGGIPMPRALQWALIEHILGANPAAYMYAMGPGMSEIFYNNGTDEQK
MLBr_02563|M.leprae_Br4923 DEALGGTPMPKALLWALHEHLMGANPAVWIYAGGAGFANILYHLGTEKQK
TH_1915|M.thermoresistible__bu EEELGGMPMPRALVWALHEHLLGANPAVWMYGGGAGFASILYKLGNEQQK
Mflv_0430|M.gilvum_PYR-GCK AEEIGGVPAPSMLEWAINELALGAQPAAFMYAAGPKMAHILHAIGNEQQR
Mvan_0238|M.vanbaalenii_PYR-1 AEEIGGVPAPAMLEWAINELALGAQPAAFMYAAGPKMADIINAIGNEEQR
Mb1502c|M.bovis_AF2122/97 AEEIGGMPAPPPLAWAVNEMIYCANPSACFFNLGPVLAQSLYIEGNDEQR
Rv1467c|M.tuberculosis_H37Rv AEEIGGMPAPPPLAWAVNEMIYCANPSACFFNLGPVLAQSLYIEGNDEQR
MMAR_2273|M.marinum_M AEEIGGMPAPPPLTWAVNEMIFCANPSAGFFILGPTMAQSLYAEGNEEQR
MUL_1869|M.ulcerans_Agy99 AEEIGGMPAPPPLTWAVNEMIFCANPSAGFFILGPTMAQSLYAEGNEEQR
MAV_3309|M.avium_104 AEEIGGMPAPPPLAWAVNEMLICANPSANFFCLGPFMSQALYLEGNEQQR
* :** * * * **: * *:*:. :: *. :: : ..::*:
MAB_4437|M.abscessus_ATCC_1997 KWAEFVAERGWGATMVLTEPDAGSDVGAGRTKAVKQDDGSWHIDGVKRFI
MSMEG_0406|M.smegmatis_MC2_155 KWATIAAERGWGATMVLTEPDAGSDVGAGRTKAVQQPDGTWHIEGVKRFI
MLBr_02563|M.leprae_Br4923 KWAVWAAERNWGSTMVLTEPDAGSDVGAGRTKAVHQEDDSWHIDGVKRFI
TH_1915|M.thermoresistible__bu KWAKIAVERQWGSTMVLTEPDAGSDVGAGRTKAIEQPDGSWHIEGVKRFI
Mflv_0430|M.gilvum_PYR-GCK HWAGLMIERAWGATMVLTEPDAGSDVGAGRTKAVQQPDGTWHLDGVKRFI
Mvan_0238|M.vanbaalenii_PYR-1 HWAALMIERNWGATMVLTEPDAGSDVGAGRTKAVQQADGTWHIDGVKRFI
Mb1502c|M.bovis_AF2122/97 RWAAEGVQRGWQATMVLTEPDAGSDVGAGRTKAFEQPDGTWHIEGVKRFI
Rv1467c|M.tuberculosis_H37Rv RWAAEGVQRGWQATMVLTEPDAGSDVGAGRTKAFEQPDGTWHIEGVKRFI
MMAR_2273|M.marinum_M RWAAMGIERGWQATMVLTEPDAGSDVGAGRAKAIEQPDGTWHIEGVKRFI
MUL_1869|M.ulcerans_Agy99 RWAAMGIERGWQATMVLTEPDAGSDVGAGRAKAIEQPDGTWHIEGVKRFI
MAV_3309|M.avium_104 RWAAEGVERGWAATMVLTEPDAGSDVGAGRAKAIEQPDGTWHIEGVKRFI
:** :* * :*****************:**..* *.:**::******
MAB_4437|M.abscessus_ATCC_1997 TSADSDDMFENIMHLVLARPEGAGPGTKGLSLFFVPKFIPNFETGEPGER
MSMEG_0406|M.smegmatis_MC2_155 TSADSDDLFENIMHLVLARPEGAGPGTKGLSLFFVPKFHFDHETGEIGER
MLBr_02563|M.leprae_Br4923 TSGDSDDLFENIFHLVLARPEGAGPGTKGLSLFFVPKFLFDFETGELGER
TH_1915|M.thermoresistible__bu TSADSDDLFENIFHLVLARPEGAGPGTKGLSLFFVPKFLFDPETGEPGER
Mflv_0430|M.gilvum_PYR-GCK TNGDADDLFENIVHVVLARPEGAGPGTKGLSLFIVPKFHFDPQTGDLGER
Mvan_0238|M.vanbaalenii_PYR-1 TNGDADDLFENILHVVLARPEGAGPGTKGLSLFLVPKFHFDPQTGEPGER
Mb1502c|M.bovis_AF2122/97 SGGDVGNTAENIFHLVLARPEGAGPGTKGLSLFYVPNYLFDPDTFELGAR
Rv1467c|M.tuberculosis_H37Rv SGGDVGNTAENIFHLVLARPEGAGPGTKGLSLFYVPNYLFDPDTFELGAR
MMAR_2273|M.marinum_M SGGDVGDTSENIFHLVLARPEGAGPGTKGLSLFYVPNYLFDPDTGELGPR
MUL_1869|M.ulcerans_Agy99 SGGDVGDTSENIFHLVLARPEGAGPGTKGLSLFYVPNYLFDPDTGELGPR
MAV_3309|M.avium_104 SGGDVGDTAENIFHLVLARPEGAGPGTKGLSLFYVPKYLFDPETFELGPR
:..* .: ***.*:****************** **:: : :* : * *
MAB_4437|M.abscessus_ATCC_1997 NGVFVTNVEHKMGLKVSATCELSFGQHGVPAKGWLVGEVHNGIAQMFDVI
MSMEG_0406|M.smegmatis_MC2_155 NGVFVTNVEHKMGLKVSATCELSLGQHGIPAVGWLVGEVHNGIAQMFDVI
MLBr_02563|M.leprae_Br4923 NGVFVTNVEHKMGLKVSATCELSFGQHGVPAKGWLVGDVHDGIAQMFEVI
TH_1915|M.thermoresistible__bu NGVFVTNVEHKMGLKVSATCELAFGQHGVPAKGWLVGDVHEGIRQMFDVI
Mflv_0430|M.gilvum_PYR-GCK NGVFVTGLEHKMGLKASATCELTFGQHGVPAVGWLVNDTHSGIAQMFKVI
Mvan_0238|M.vanbaalenii_PYR-1 NGVFVTGLEHKMGLKASATCELTFGQHGVPAVGWLVNDTHNGIAQMFKVI
Mb1502c|M.bovis_AF2122/97 NGVYVTGLEHKMGLKSSPTCELTFGGADVPAVGYLVGGVHNGIAQMFTVI
Rv1467c|M.tuberculosis_H37Rv NGVYVTGLEHKMGLKSSPTCELTFGGADVPAVGYLVGGVHNGIAQMFTVI
MMAR_2273|M.marinum_M NGVYVTGLEHKMGLKSSPTCEVTFGATDVPAVGYLVGGVHNGIAQMFTVI
MUL_1869|M.ulcerans_Agy99 NGVYVTGLEHKMGLKSSPTCEVTFGATDVPAVGYLVGGVHNGIAQMFTVI
MAV_3309|M.avium_104 NGVFVTGVEHKMGIKSSPTCELTFGATDVPAVGYLVGDVHRGIAQMFTVI
***:**.:*****:* *.***:::* .:** *:**. .* ** *** **
MAB_4437|M.abscessus_ATCC_1997 EQARMMVGTKAIATLSTGYLNALEYAKTRVQGADMTQLTDKTAPRVTITH
MSMEG_0406|M.smegmatis_MC2_155 EQARMMVGTKAIATLSTGYLNALEYAKERVQGADMTQMTDKTAPRVTITH
MLBr_02563|M.leprae_Br4923 EQARMMVGTKAIATLSTGYLNALEYAKSRVQGADLTQMTDKTAPRVTITH
TH_1915|M.thermoresistible__bu EQARMMVGTKAIATLSTGYLNALEYAKERVQGADMTQMTDKTAPRVTITH
Mflv_0430|M.gilvum_PYR-GCK EYARMMVGTKAIATLSTGYLNALAYAKTRIQGADMIQMTDKTAPRVTIMH
Mvan_0238|M.vanbaalenii_PYR-1 EYARMMVGTKAMSTLSTGYLNALEYAKTRVQGADMTQMTDKSAPRVTIIH
Mb1502c|M.bovis_AF2122/97 EHARMTIGVKSAGTLSTGYLNALAFAKERVQGADLTQMTDKTAPRVTIMH
Rv1467c|M.tuberculosis_H37Rv EHARMTIGVKSAGTLSTGYLNALAFAKERVQGADLTQMTDKTAPRVTIMH
MMAR_2273|M.marinum_M EHARMTIGVKSSGTLSTGYLNALAYAKERVQGADLTQMTDKTAPRVTIMH
MUL_1869|M.ulcerans_Agy99 EHARMTIGVKSSGTLSTGYLNALAYAKERVQGADLTQMTDKTAPRVTIMH
MAV_3309|M.avium_104 EQARMTIGVKAAGTLSTGYLNALAFAKERVQGADLTQMADKTAPRVTIIH
* *** :*.*: .********** :** *:****: *::**:****** *
MAB_4437|M.abscessus_ATCC_1997 HPDVRRSLMTQKVYAEGLRALYLYATTFQDAVAAKTINGVEAEDAVKLND
MSMEG_0406|M.smegmatis_MC2_155 HPDVRRSLMTQKAYAEGLRAIYLYTATFQDAEVAQAVHGVDGDLAARVND
MLBr_02563|M.leprae_Br4923 HPDVRRSLMTQKAYAEGLRALYLYTATFQDAAVAEAVHGVDAKLAVKIND
TH_1915|M.thermoresistible__bu HPDVRRSLMTQKAYAEGMRAMYLYTATYQDAAVAEAVHGVDADLAMRVND
Mflv_0430|M.gilvum_PYR-GCK HPDVRRALMTQKAYAEGLRALYLYTAAHQDTTVADIVSGADAAMAGRVND
Mvan_0238|M.vanbaalenii_PYR-1 HPDVRRALLIQKAYAEGLRALYLYTAAHQDPAAADIVSGADAAMADRVND
Mb1502c|M.bovis_AF2122/97 HPDVRRSLMTQKAYAEGLRALYLYAAAHQDDAVAQRVSGADHDMAHRVDD
Rv1467c|M.tuberculosis_H37Rv HPDVRRSLMTQKAYAEGLRALYLYAAAHQDDAVAQRVSGADHDMAHRVDD
MMAR_2273|M.marinum_M HPDVRRSLMTQKAYAEGLRALYLYAAAHQDDAVAQRVSGADHDMAHRVDD
MUL_1869|M.ulcerans_Agy99 HPDVRRSLMTQKAYAEGLRALYLYAAAHQDDAVAQRVSGADHDMAHRVDD
MAV_3309|M.avium_104 HPDVRRSLMTQKAYAEGLRALYMYAAAHQDDQVAQRVSGADHDMAHRVDD
******:*: **.****:**:*:*:::.** .*. : *.: * :::*
MAB_4437|M.abscessus_ATCC_1997 LLLPVIKGVGSERAYEKLTESLQTFGGSGFLQDYPIEQYIRDAKIDSLYE
MSMEG_0406|M.smegmatis_MC2_155 LLLPIVKGFGSETAYAKLTESLQTLGGSGFLQDYPIEQYIRDSKIDSLYE
MLBr_02563|M.leprae_Br4923 LMLPLVKGVGSEQAYAKLTESLQTLGGSGFLQDYPIEQYIRDAKIDSLYE
TH_1915|M.thermoresistible__bu LLLPIVKGVGSEQAYAKLTESLQTLGGSGFLQDYPIEQYIRDSKIDSLYE
Mflv_0430|M.gilvum_PYR-GCK LLLPIVKGVGSERAYQCLTESLQTFGGSGFLQDYPVEQYIRDAKIDSLYE
Mvan_0238|M.vanbaalenii_PYR-1 LLLPIVKGVGSERAYQCLTESLQTFGGSGFLQDYPIEQYIRDAKIDSLYE
Mb1502c|M.bovis_AF2122/97 LLLPIVKGVGSERAYEILTESLQTLGGSGFLVDYPLEQYIRDAKIDSLYE
Rv1467c|M.tuberculosis_H37Rv LLLPIVKGVGSERAYEILTESLQTLGGSGFLVDYPLEQYIRDAKIDSLYE
MMAR_2273|M.marinum_M LLLPVVKGVGSERAYEILTESLQTLGGSGFLSDYPIEQYIRDAKIDSLYE
MUL_1869|M.ulcerans_Agy99 LLLPVVKGVGSERAYEILTESLQTLGGSGFLSDYPIEQYIRDAKIDSLYE
MAV_3309|M.avium_104 LLLPIVKGVSSERAYEVLTESLQTLGGSGFLQDYPLEQYIRDSKIDSLYE
*:**::**..** ** *******:****** ***:******:*******
MAB_4437|M.abscessus_ATCC_1997 GTTAIQAQDFFFRKIVKDQGKSLAFISGQIEEFVKSETGNGRLKAERALL
MSMEG_0406|M.smegmatis_MC2_155 GTTAIQAQDFFFRKIIRDKGQALAYVAGEIEQFIKNENGNGRLKTERELL
MLBr_02563|M.leprae_Br4923 GTTAIQAQDFFFRKIVRDKGVALSYVSGQIQQFVDSETGNGRLKSERELL
TH_1915|M.thermoresistible__bu GTTAIQAQDFFFRKIARDQGVALTHVVGQIQAFIDSDTARPELADERALL
Mflv_0430|M.gilvum_PYR-GCK GTTAIQAQDFFFRKIARDQGGALLHVVSHMRRFLDDAAARPELATERALL
Mvan_0238|M.vanbaalenii_PYR-1 GTTAIQAQDFFFRKIARDQGGALLHVVSQLRRFLDDLSAREELATERALL
Mb1502c|M.bovis_AF2122/97 GTTAIQALDFFFRKIVRDHGKALQFVLAQVTHTVEN--IDPSLKPQAELL
Rv1467c|M.tuberculosis_H37Rv GTTAIQALDFFFRKIVRDHGKALQFVLAQVTHTVEN--IDPSLKPQAELL
MMAR_2273|M.marinum_M GTTAIQSLDFFFRKIVRDHGQALQFVTAQITQTIDN--VDPSLKPQAELL
MUL_1869|M.ulcerans_Agy99 GTTAIQSLDFFFRKIVRDHGQALQFVTAQITQTIDN--VDPSLKPQAELL
MAV_3309|M.avium_104 GTTAIQALDFFFRKIVRDRGQALQFVTAQITATVDD--CDEALRPIAELL
******: ******* :*:* :* .: ..: :.. * **
MAB_4437|M.abscessus_ATCC_1997 ATALEDVQGMAASMTGNLMAAQQEITQVYRVGTASVRFLMSVGDLLIGWL
MSMEG_0406|M.smegmatis_MC2_155 ATALADVQGMAASLTGYLMAAQEDAASIYKVGLGSVRFLMAVGDLLSGWL
MLBr_02563|M.leprae_Br4923 AKALTDIQGMEASLTGYLMAAQQDVTSLYKVGLGSVRFLMSVGDLIIGWL
TH_1915|M.thermoresistible__bu AAALSDAQAMVAALTGYLMKSQETPEELYRIGLESVRFLLAAGDLLIGWL
Mflv_0430|M.gilvum_PYR-GCK ATAVGDVQEMVAAMTKFLVDSQENPRELYRLGLESVPFLLSVGDLLLAWL
Mvan_0238|M.vanbaalenii_PYR-1 GTAVDDVQEMVAAMTKYLVDSQQNARELYRLGLESVPFLLAVGDLLLGWL
Mb1502c|M.bovis_AF2122/97 RTALDDITAMTGALTGYLMSAAQHSSDIYKVGLGSVRYLLAVGDLLIGWR
Rv1467c|M.tuberculosis_H37Rv RTALDDITAMTGALTGYLMSAAQHSSDIYKVGLGSVRYLLAVGDLLIGWR
MMAR_2273|M.marinum_M QTALDDVTAMTGTLTTYLMSAAQHPTDIYKVGLGSVRYLLAVGDLIIGWR
MUL_1869|M.ulcerans_Agy99 QTALDDVTAMTGTLTTYLMSAAQHPTDIYKVGLGSVRYLLAVGDLIIGWR
MAV_3309|M.avium_104 QTAFDDVTAMTGALTGYLMSAAEQPTDIYKVGLASVRYLLAVGDLLIGWR
*. * * .::* *: : : .:*::* ** :*::.***: .*
MAB_4437|M.abscessus_ATCC_1997 LQRQAAVAIEALDAGATGADKSFYEGKIAAASFFAKNMLPLLTSTRAVLE
MSMEG_0406|M.smegmatis_MC2_155 LARQAAVAIEKLDAGATGADKSFYEGKIAAASFFAKNMLPLLTSTRQIIE
MLBr_02563|M.leprae_Br4923 LQRQAAVAVQALDAGASSAERSFYEGKVAVSSFFAKNFLPLLTSTREVLE
TH_1915|M.thermoresistible__bu LLWHAEIALAALDGDVADAERHFYVGKVAAARFFARNVLPRLSAERTMVE
Mflv_0430|M.gilvum_PYR-GCK LLQQAEIALNALDGTVSERDRAFYTGKVATGKFFARNVLPRLSAQRAILQ
Mvan_0238|M.vanbaalenii_PYR-1 LLHQAETALTALEGDVSERDRAFYTGKVATAKFFARNVLPRLTAQRGILQ
Mb1502c|M.bovis_AF2122/97 LLVLAGVAHAALADGPSQNDEAFYRGKIAVAAFFAKNMLPKLTGVRSVIE
Rv1467c|M.tuberculosis_H37Rv LLVLAGVAHAALADGPSQNDEAFYRGKIAVAAFFAKNMLPKLTGVRSVIE
MMAR_2273|M.marinum_M LLVQAGVAHAALADSPSKDDEAFYKGKIAVATFFAKNMLPKLTGVRAVIE
MUL_1869|M.ulcerans_Agy99 LLVQAGVAHAALADSPSKDDEAFYKGKIAVATFFAKNMLPKLTGVRAVIE
MAV_3309|M.avium_104 LLVHADVAQRALAAGAGGDDEAFYRGKIATAAFFAKNMLPRLSGLRGIIE
* * * * :. ** **:*.. ***:*.** *:. * :::
MAB_4437|M.abscessus_ATCC_1997 NVDNDIMELDEAAF
MSMEG_0406|M.smegmatis_MC2_155 NLDNDVMELDEAAF
MLBr_02563|M.leprae_Br4923 VLDNDIMELDEAAF
TH_1915|M.thermoresistible__bu SVDRSIMELPEAAF
Mflv_0430|M.gilvum_PYR-GCK AVDLTAMDLPEAAF
Mvan_0238|M.vanbaalenii_PYR-1 AVDLTAMELSEAGF
Mb1502c|M.bovis_AF2122/97 NIDDDIMRVPEDAF
Rv1467c|M.tuberculosis_H37Rv NIDDDIMRVPEDAF
MMAR_2273|M.marinum_M SIDDEIMRISEEAF
MUL_1869|M.ulcerans_Agy99 SIDDEIMRISEEAF
MAV_3309|M.avium_104 AIDDEIMKLPEAAF
:* * : * .*