For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAVQAARRARIERRTKESDIVIELDLDGTGRVDVETGVPFYDHMLTALGSHASFDLTVRTTGDVEIEAHH TIEDTAIALGAALGQALGDKRGIRRFGDAFIPMDETLAHAAVDVSGRPYCVHSGEPDHLQHSTIAGSSVP YHTVINRHVFESLAMNARIALHVRVLYGRDPHHITEAQYKAVARALRQAVEPDPRVSDVPSTKGVL*
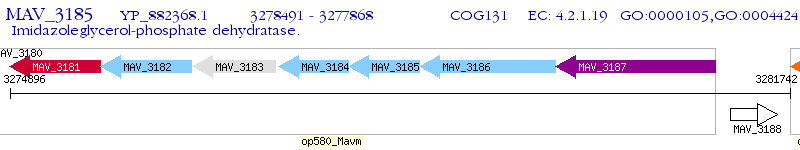
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3185 | hisB | - | 100% (207) | imidazoleglycerol-phosphate dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1627 | hisB | 1e-108 | 93.10% (203) | imidazoleglycerol-phosphate dehydratase |
| M. gilvum PYR-GCK | Mflv_3611 | hisB | 2e-93 | 79.60% (201) | imidazoleglycerol-phosphate dehydratase |
| M. tuberculosis H37Rv | Rv1601 | hisB | 1e-108 | 93.10% (203) | imidazoleglycerol-phosphate dehydratase |
| M. leprae Br4923 | MLBr_01259 | hisB | 2e-98 | 85.22% (203) | imidazoleglycerol-phosphate dehydratase |
| M. abscessus ATCC 19977 | MAB_2668c | hisB | 2e-98 | 81.64% (207) | imidazoleglycerol-phosphate dehydratase |
| M. marinum M | MMAR_2397 | hisB | 1e-108 | 90.95% (210) | imidazole glycerol-phosphate dehydratase HisB |
| M. smegmatis MC2 155 | MSMEG_3207 | hisB | 2e-93 | 80.69% (202) | imidazoleglycerol-phosphate dehydratase |
| M. thermoresistible (build 8) | TH_1161 | hisB | 4e-96 | 83.58% (201) | Probable imidazole glycerol-phosphate dehydratase hisB |
| M. ulcerans Agy99 | MUL_1574 | hisB | 1e-108 | 90.95% (210) | imidazoleglycerol-phosphate dehydratase |
| M. vanbaalenii PYR-1 | Mvan_2806 | hisB | 2e-97 | 81.86% (204) | imidazoleglycerol-phosphate dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1627|M.bovis_AF2122/97 MTTTQTAKASRRARIERRTRESDIVIELDLDGTGQVAVDTGVPFYDHMLT
Rv1601|M.tuberculosis_H37Rv MTTTQTAKASRRARIERRTRESDIVIELDLDGTGQVAVDTGVPFYDHMLT
MAV_3185|M.avium_104 MTAVQ---AARRARIERRTKESDIVIELDLDGTGRVDVETGVPFYDHMLT
MMAR_2397|M.marinum_M MTATETGTAVRRARIERRTRESDIVIDLDLDGTGQVDVDTGVPFFDHMLT
MUL_1574|M.ulcerans_Agy99 MTATETGTAVRRARIERRTRESDIVIDLDLDGTGQVDVDTGVPFFDHMLT
MLBr_01259|M.leprae_Br4923 MTNTEVGKTTRRARIERRTSESDIVVELDLDGTGQVHIDTGVSFYDHMLT
MAB_2668c|M.abscessus_ATCC_199 MTVPT---QARTARVARATRESDIVVELNLDGSGQVQIDTGVPFYDHMLT
Mflv_3611|M.gilvum_PYR-GCK --MS------RFAKVERKTKESDIVVELDLDGTGQVSIDTGVPFFDHMLT
Mvan_2806|M.vanbaalenii_PYR-1 --MSDPIRPARRAKVERKTKESDIVVELDLDGTGQVSVDTGVPFFDHMLT
MSMEG_3207|M.smegmatis_MC2_155 --MSALAN--RRARVERKTKESEIVVDLDLDGTGVVDIDTGVPFFDHMLT
TH_1161|M.thermoresistible__bu --VTDILTGARRARVERTTTESEIVVELDLDGTGVVDVSTGVPFFDHMLT
* *:: * * **:**::*:***:* * :.***.*:*****
Mb1627|M.bovis_AF2122/97 ALGSHASFDLTVRATGDVEIEAHHTIEDTAIALGTALGQALGDKRGIRRF
Rv1601|M.tuberculosis_H37Rv ALGSHASFDLTVRATGDVEIEAHHTIEDTAIALGTALGQALGDKRGIRRF
MAV_3185|M.avium_104 ALGSHASFDLTVRTTGDVEIEAHHTIEDTAIALGAALGQALGDKRGIRRF
MMAR_2397|M.marinum_M ALGSHASFDLTVRTKGDVEIEAHHTVEDTAIALGQALGQALGDKKGIRRF
MUL_1574|M.ulcerans_Agy99 ALGSHASFDLTVRTKGDVEIEAHHTVEDTAIALGQALGQALGDKKGIRRF
MLBr_01259|M.leprae_Br4923 ALGSHASFDLTVCTKGDVEIEAHHTIEDTAIALGQAFGQALGNKKGIRRF
MAB_2668c|M.abscessus_ATCC_199 ALGTHASFDLTITAKGDTEIEAHHTVEDTAITLGQALAEALGDKKGIRRF
Mflv_3611|M.gilvum_PYR-GCK SLGTHASFDLTVKAVGDIEIEGHHTIEDTSIVLGQALAQALGDKKGIRRF
Mvan_2806|M.vanbaalenii_PYR-1 SLGSHASFDLTIKAVGDIEIEGHHTIEDTAIVLGQALGQALGDKKGIRRF
MSMEG_3207|M.smegmatis_MC2_155 SLGSHASFDLTVHAKGDIEIEGHHTVEDTAIVLGQALGQALGDKKGIRRF
TH_1161|M.thermoresistible__bu ALGSHASFDLTVRARGDIDIEGHHTIEDTAIVLGQALGQALGDKKGIRRF
:**:*******: : ** :**.***:***:*.** *:.:***:*:*****
Mb1627|M.bovis_AF2122/97 GDAFIPMDETLAHAAVDLSGRPYCVHTGEPDHLQHTTIAGSSVPYHTVIN
Rv1601|M.tuberculosis_H37Rv GDAFIPMDETLAHAAVDLSGRPYCVHTGEPDHLQHTTIAGSSVPYHTVIN
MAV_3185|M.avium_104 GDAFIPMDETLAHAAVDVSGRPYCVHSGEPDHLQHSTIAGSSVPYHTVIN
MMAR_2397|M.marinum_M GDAFIPMDETLAHAAVDVSGRPYCVHTGEPDHLQHTTIAGNSVPYHTVIN
MUL_1574|M.ulcerans_Agy99 GDAFIPMDETLAHAAVDVSGRPYCVHTGEPDHLQHTTIAGNSVPYHTVIN
MLBr_01259|M.leprae_Br4923 GDAFIPMDETLVHAVVDVSGRPYCVHTGEPDHLQHNIISGSSVPYSTVIN
MAB_2668c|M.abscessus_ATCC_199 GDAFIPMDETLAHAVVDVSGRPYCVHTGEPDHLLHTTIAGSSVPYHTVIN
Mflv_3611|M.gilvum_PYR-GCK GDAFIPMDECLAHAAVDVSGRPYFVHTGEPDYMVQFTIAGSAAPYHTVVN
Mvan_2806|M.vanbaalenii_PYR-1 GDAFIPMDETLAHAAVDVSGRPYFVHTGEPDYMVEFTIAGSAAPYHTVVN
MSMEG_3207|M.smegmatis_MC2_155 GDAFIPMDESLAHAAVDVSGRPYFVHTGEPESMVSFTIAGTGAPYHTVIN
TH_1161|M.thermoresistible__bu GDAFIPMDEALAHAAVDLSGRPYFVHTGEPEYMVEFTIAGSAAPYHTVIN
********* *.**.**:***** **:***: : *:*...** **:*
Mb1627|M.bovis_AF2122/97 RHVFESLAANARIALHVRVLYGRDPHHITEAQYKAVARALRQAVEPDPRV
Rv1601|M.tuberculosis_H37Rv RHVFESLAANARIALHVRVLYGRDPHHITEAQYKAVARALRQAVEPDPRV
MAV_3185|M.avium_104 RHVFESLAMNARIALHVRVLYGRDPHHITEAQYKAVARALRQAVEPDPRV
MMAR_2397|M.marinum_M RHVFESLAMNARIALHVRVLYGRDPHHITEAQYKAVARALRQAVEPDPRV
MUL_1574|M.ulcerans_Agy99 RHVFESLAMNARIALHVRVLYGRDPHHITEAQYKAVARALRQAVEPDPRV
MLBr_01259|M.leprae_Br4923 RHVFESLAANARIALHVRVLYGRDPHHITEAQYKAVARALSEAVKFDPRF
MAB_2668c|M.abscessus_ATCC_199 AHVFESLALNARIALHVRTLYGRDPHHITEAQYKAVARALRQAVEPDPRV
Mflv_3611|M.gilvum_PYR-GCK RHVFESLAYNARIALHVRTLYGRDPHHITEAEYKAVARALRQAVEYDPRV
Mvan_2806|M.vanbaalenii_PYR-1 RHVFESLAFNARIALHVRTLYGRDPHHITEAEYKAVARALRQAVEYDPRV
MSMEG_3207|M.smegmatis_MC2_155 RHVFESLAFNARIALHVRTLYGRDPHHITEAQYKAVARALRQAVEYDARV
TH_1161|M.thermoresistible__bu RHVFESLAFNARIALHVRTLYGRDPHHITEAQYKAVARALRHAVEPDPRV
******* *********.************:******** .**: *.*.
Mb1627|M.bovis_AF2122/97 SGVPSTKGAL
Rv1601|M.tuberculosis_H37Rv SGVPSTKGAL
MAV_3185|M.avium_104 SDVPSTKGVL
MMAR_2397|M.marinum_M SGVPSTKGVL
MUL_1574|M.ulcerans_Agy99 SGVPSTKGVL
MLBr_01259|M.leprae_Br4923 SGVPSTKGVL
MAB_2668c|M.abscessus_ATCC_199 TGVPSTKGSL
Mflv_3611|M.gilvum_PYR-GCK TGVPSTKGTL
Mvan_2806|M.vanbaalenii_PYR-1 SGVPSTKGTL
MSMEG_3207|M.smegmatis_MC2_155 TGVPSTKGTL
TH_1161|M.thermoresistible__bu SGVPSTKGAL
:.****** *