For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HAHLAATTSREDFRQLAAEHRVVPVTRKVLADSETPLSAYRKLAANRPGTFLLESAEHGRSWSRWSFIGA GSPSALTVRDGEAGWLGVVPQDAPTGGDPLQALRATLDLLATGPLSGLPPLSGGMVGFFAYDLVRRLERL PELAVDDLGLPDMLLLLATDLAAVDHHEGTITLIANAVNWNGTDERVDEAYDDAVARLDVMTAALGQPLA STVATFERPEPRYRAQRSVEEYGKIVDYLVEQIAAGEAFQVVPSQRFEMDTDVDPIDVYRMLRVTNPSPY MYLLHVPNDAGGTDFSIVGSSPEALVTVADGVATTHPIAGTRWRGQTDEEDQLLEKELLADEKERAEHLM LVDLGRNDLGRVCAPGTVRVDDYSHIERYSHVMHLVSTVTGVLDAGRTALDAVTACFPAGTLSGAPKVRA MELIEEVEKTRRGLYGGVVGYLDFAGNADFAIAIRTALMRGGTAYVQSGGGVVADSNGPYEYTEATNKAR AVLGAIAAAETLTEPGARR*
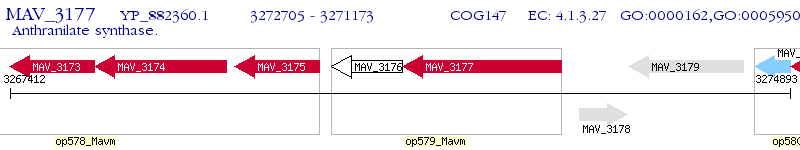
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3177 | trpE | - | 100% (510) | anthranilate synthase component I |
| M. avium 104 | MAV_1792 | - | 9e-31 | 33.20% (250) | salicylate synthase MbtI |
| M. avium 104 | MAV_1127 | pabB | 5e-27 | 32.59% (270) | para-aminobenzoate synthase component I |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1635 | trpE | 0.0 | 88.30% (513) | anthranilate synthase component I |
| M. gilvum PYR-GCK | Mflv_3602 | - | 0.0 | 87.50% (504) | anthranilate synthase component I |
| M. tuberculosis H37Rv | Rv1609 | trpE | 0.0 | 88.30% (513) | anthranilate synthase component I |
| M. leprae Br4923 | MLBr_01269 | trpE | 0.0 | 88.10% (504) | anthranilate synthase component I |
| M. abscessus ATCC 19977 | MAB_2647c | - | 0.0 | 82.77% (499) | anthranilate synthase component I |
| M. marinum M | MMAR_2411 | trpE | 0.0 | 86.22% (508) | anthranilate synthase component I TrpE |
| M. smegmatis MC2 155 | MSMEG_3217 | trpE | 0.0 | 86.51% (504) | anthranilate synthase component I |
| M. thermoresistible (build 8) | TH_2725 | - | 0.0 | 88.61% (404) | PUTATIVE anthranilate synthase component I |
| M. ulcerans Agy99 | MUL_1589 | trpE | 0.0 | 86.02% (508) | anthranilate synthase component I |
| M. vanbaalenii PYR-1 | Mvan_2814 | - | 0.0 | 87.90% (504) | anthranilate synthase component I |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3602|M.gilvum_PYR-GCK MQTTAD----------------LSVTTSREDFRTLAAEHRVVPVTRKVLA
Mvan_2814|M.vanbaalenii_PYR-1 MQMTAT----------------LSATTTREDFRALAAEHRVVPVTRKVLA
MSMEG_3217|M.smegmatis_MC2_155 MQTTANHSSRSTQTGTRAHGAALAETTSREDFRALATEHRVVPVIRKVLA
TH_2725|M.thermoresistible__bu --------------------------------------------------
Mb1635|M.bovis_AF2122/97 --MHAD----------------LAATTSREDFRLLAAEHRVVPVTRKVLA
Rv1609|M.tuberculosis_H37Rv --MHAD----------------LAATTSREDFRLLAAEHRVVPVTRKVLA
MMAR_2411|M.marinum_M --MRAD----------------LATTITRENFRSLAAEHRVVPVTRKVLA
MUL_1589|M.ulcerans_Agy99 --MRAD----------------LATTITRENFRSLAAERRVVPVTRKVLA
MLBr_01269|M.leprae_Br4923 --MHAH----------------LAATTSREDFRQLAVDHRVVPVTRKVLA
MAV_3177|M.avium_104 --MHAH----------------LAATTSREDFRQLAAEHRVVPVTRKVLA
MAB_2647c|M.abscessus_ATCC_199 --MHSTVSG--------------AATTTREEFNALAAVHRVVPVTRKVLA
Mflv_3602|M.gilvum_PYR-GCK DSETPLSAYRKLAANRPGTFLLESAENGRSWSRWSFIGAGAPSALTVRDG
Mvan_2814|M.vanbaalenii_PYR-1 DSETPLSAYRKLAANRPGTFLLESAENGRSWSRWSFIGAGAPSALTVRDG
MSMEG_3217|M.smegmatis_MC2_155 DSETPLSAYRKLAANRPGTFLLESAENGRSWSRWSFIGAGAPSALTVRDN
TH_2725|M.thermoresistible__bu ----------------------------------------------VXX-
Mb1635|M.bovis_AF2122/97 DSETPLSAYRKLAANRPGTFLLESAENGRSWSRWSFIGAGAPTALTVREG
Rv1609|M.tuberculosis_H37Rv DSETPLSAYRKLAANRPGTFLLESAENGRSWSRWSFIGAGAPTALTVREG
MMAR_2411|M.marinum_M DSETPLSAYRKLADGRPGTFLLESAENGRSWSRWSFIGAGAPTALTVRDG
MUL_1589|M.ulcerans_Agy99 DSETPLSAYRKLADGRPGTFLLESAENGRSWSRWSFIGAGAPTALTVRDG
MLBr_01269|M.leprae_Br4923 DSETPLSAYRKLAANRPSTFLLESAENGRSWSQWSFIGVGAPSALTIRDG
MAV_3177|M.avium_104 DSETPLSAYRKLAANRPGTFLLESAEHGRSWSRWSFIGAGSPSALTVRDG
MAB_2647c|M.abscessus_ATCC_199 DSETPLSAYRKLGANRPGTFLLESAENGRSWSRWSFIGAGSPSALTVRDG
:
Mflv_3602|M.gilvum_PYR-GCK EAVWLGVIPQDAPAGGDPLQALRTTLTLLETA---ALPGLPPLSSGLVGF
Mvan_2814|M.vanbaalenii_PYR-1 EAVWLGVIPQDAPSGGDPLQALRATLTLLETA---PLPGLPPLSSGLVGF
MSMEG_3217|M.smegmatis_MC2_155 AAAWLGTAPEGAPSGGDPLDALRATLDLLKTE---AMAGLPPLSSGLVGF
TH_2725|M.thermoresistible__bu -------------------XALRATLELLKTE---PLPGLPPLSSGLVGF
Mb1635|M.bovis_AF2122/97 QAVWLGAVPKDAPTGGDPLRALQVTLELLATADRQSEPGLPPLSGGMVGF
Rv1609|M.tuberculosis_H37Rv QAVWLGAVPKDAPTGGDPLRALQVTLELLATADRQSEPGLPPLSGGMVGF
MMAR_2411|M.marinum_M EAVWLGTAPEGAPTGGDPLRALQSTLEVLATG---VMPGLPPLSGGMVGF
MUL_1589|M.ulcerans_Agy99 EAVWLGTAPEGAPTGGDPLRALQSTLEVLATG---VMPGLPPLSGGMVGF
MLBr_01269|M.leprae_Br4923 EAVWLGTVPQDAPTGGDPLHVLQATLELLATA---AMPGLPPLSSGMVGF
MAV_3177|M.avium_104 EAGWLGVVPQDAPTGGDPLQALRATLDLLATG---PLSGLPPLSGGMVGF
MAB_2647c|M.abscessus_ATCC_199 QSAWLGATPEGAPVGGDPLDALRATMDLLAGE---SLAGLPPLSGGMVGF
.*: *: :* .******.*:***
Mflv_3602|M.gilvum_PYR-GCK FAYDLVRRLERLPELTVDDLGLPDMMLLLATDIAAVDHHEGTITLIANAV
Mvan_2814|M.vanbaalenii_PYR-1 FAYDLVRRLERLPELTVDDLALPDMMLLLATDVAAVDHHEGTITLIANAV
MSMEG_3217|M.smegmatis_MC2_155 FAYDMVRRLERLPELAVDDLGLPDMLLLLATDIAAVDHHEGTITLIANAV
TH_2725|M.thermoresistible__bu FAYDLVRRLERLPELAVDDLGLPDMVLLLATDIAAVDHHEGTITLIANAV
Mb1635|M.bovis_AF2122/97 FAYDMVRRLERLPERAVDDLCLPDMLLLLATDVAAVDHHEGTITLIANAV
Rv1609|M.tuberculosis_H37Rv FAYDMVRRLERLPERAVDDLCLPDMLLLLATDVAAVDHHEGTITLIANAV
MMAR_2411|M.marinum_M FAYDMVRRLERLPNLAVDDLQLPEMLLLLATDLAAVDHHEGTITLIANAV
MUL_1589|M.ulcerans_Agy99 FAYDMVRRLERLPNLAVDDLHLPEMLLLLATDLAAVDHHEGTITLIANAV
MLBr_01269|M.leprae_Br4923 FAYDMVRRLERLPELALNDLQLPDMLLLLATDVAAVDHHEGTITLIANAV
MAV_3177|M.avium_104 FAYDLVRRLERLPELAVDDLGLPDMLLLLATDLAAVDHHEGTITLIANAV
MAB_2647c|M.abscessus_ATCC_199 LAYDIVRRLEKLPELAADDLGLPDLLLLLATDMAAVDHHEGTITLIANAV
:***:*****:**: : :** **:::******:*****************
Mflv_3602|M.gilvum_PYR-GCK NWNGTDERVDEAYDDAVARLDVMTAALAEPLASTVATFRRPAPQPRAQLS
Mvan_2814|M.vanbaalenii_PYR-1 NWNGTDERVDWAYDDAVARLDVMTAALAEPLASTVATFSRPVPEHRSQRT
MSMEG_3217|M.smegmatis_MC2_155 NWNGTDERVDWAYDDAVARLDVMTKALGQPLTSAVATFSRPAPDHRAQRT
TH_2725|M.thermoresistible__bu NWNGTDERVDWAYDDAVARLDVMTAALGEPLPSAVATFSRPEPRYRAQRT
Mb1635|M.bovis_AF2122/97 NWNGTDERVDWAYDDAVARLDVMTAALGQPLPSTVATFSRPEPRHRAQRT
Rv1609|M.tuberculosis_H37Rv NWNGTDERVDWAYDDAVARLDVMTAALGQPLPSTVATFSRPEPRHRAQRT
MMAR_2411|M.marinum_M NWDGTDERVDWAYDDAIARLDVMTAALSQPLPSSVATFSRPEPHYRAQRT
MUL_1589|M.ulcerans_Agy99 NWDGTDERVDWAYDDAIARLDVMTAALSQPLPSSVATFSRPEPHYRAQRT
MLBr_01269|M.leprae_Br4923 NWNGTDERVDQAYDDAIARLDVMTAALGQPLPSTIATFSRPDPRRRAQCT
MAV_3177|M.avium_104 NWNGTDERVDEAYDDAVARLDVMTAALGQPLASTVATFERPEPRYRAQRS
MAB_2647c|M.abscessus_ATCC_199 NWDDTPERVAEAYDGAVARLDVMTAALGQPLPSTVAHFDRPAPTYRSQRT
**:.* *** ***.*:******* **.:**.*::* * ** * *:* :
Mflv_3602|M.gilvum_PYR-GCK VDEYTAIVDKLVGDIEAGEAFQVVPSQRFEMDTAADPLDVYRMLRVTNPS
Mvan_2814|M.vanbaalenii_PYR-1 VEEYTAIVDKLVGDIEAGEAFQVVPSQRFEMDTDADPLDVYRMLRVTNPS
MSMEG_3217|M.smegmatis_MC2_155 MEEYTEIVDKLVGDIEAGEAFQVVPSQRFEMDTAADPLDVYRILRVTNPS
TH_2725|M.thermoresistible__bu VEEYTAIVEKLVGDIEAGEAFQVVPSQRFEMDTDADPLDVYRMLRVSNPS
Mb1635|M.bovis_AF2122/97 VEEYGAIVEYLVDQIAAGEAFQVVPSQRFEMDTDVDPIDVYRILRVTNPS
Rv1609|M.tuberculosis_H37Rv VEEYGAIVEYLVDQIAAGEAFQVVPSQRFEMDTDVDPIDVYRILRVTNPS
MMAR_2411|M.marinum_M VEEYGAIVEHLVEEIAAGEAFQVVPSQRFEMDTTVDPIDVYRILRVTNPS
MUL_1589|M.ulcerans_Agy99 VEEYGAIVEHLVEEIAAGEAFQVVPSQRFEMDTTVDPIDVYRILRVTNPS
MLBr_01269|M.leprae_Br4923 IEEYGAIVDHLVDQIAAGEAFQVVPSQRFEVDTDVDPIDVYRMLRVTNPS
MAV_3177|M.avium_104 VEEYGKIVDYLVEQIAAGEAFQVVPSQRFEMDTDVDPIDVYRMLRVTNPS
MAB_2647c|M.abscessus_ATCC_199 VQEYSAIVDKLVGDIEAGEAFQVVPSMRFEMETDVDPLEVYRILRVTNPS
::** **: ** :* ********** ***::* .**::***:***:***
Mflv_3602|M.gilvum_PYR-GCK PYMYLLNVPDHDGGLDFSIVGSSPEALVTVKDGRATTHPIAGTRWRGDTE
Mvan_2814|M.vanbaalenii_PYR-1 PYMYLLNVPNADGGLDFSIVGSSPEALVTVKDGRATTHPIAGTRWRGDTE
MSMEG_3217|M.smegmatis_MC2_155 PYMYLLNIPDADGGLDFSIVGSSPEALVTVKDGRATTHPIAGTRWRGATE
TH_2725|M.thermoresistible__bu PYMYLLHVPDDAGGLDFSIVGSSPEALVTVKDGRATTHPIAGTRWRGATE
Mb1635|M.bovis_AF2122/97 PYMYLLQVPNSDGAVDFSIVGSSPEALVTVHEGWATTHPIAGTRWRGRTD
Rv1609|M.tuberculosis_H37Rv PYMYLLQVPNSDGAVDFSIVGSSPEALVTVHEGWATTHPIAGTRWRGRTD
MMAR_2411|M.marinum_M PYMYLIQVPNDHGSVEFSIVGSSPEALVTVHDGCATTHPIAGTRWRGETD
MUL_1589|M.ulcerans_Agy99 PYMYLIQVPNDHGSVEFSIVGSSPEALVTVHDGCATTHPIAGTRWRGETD
MLBr_01269|M.leprae_Br4923 PYMYLLHVPNSDGATGFSIVGSSPEALVTVKDGRVTTHPIAGTRWRGQTE
MAV_3177|M.avium_104 PYMYLLHVPNDAGGTDFSIVGSSPEALVTVADGVATTHPIAGTRWRGQTD
MAB_2647c|M.abscessus_ATCC_199 PYMYLLHAPDSEGGTAFSIVGSSPEALVTVQDGVATTHPIAGTRWRGATD
*****:: *: *. ************** :* .************ *:
Mflv_3602|M.gilvum_PYR-GCK EEDILLEKELLSDEKERAEHLMLVDLGRNDLGRVCRPGTVKVEDYSHIER
Mvan_2814|M.vanbaalenii_PYR-1 EEDLLLEKELLCDEKERAEHLMLVDLGRNDLGRVCRPGTVKVEDYSHIER
MSMEG_3217|M.smegmatis_MC2_155 EEDVLLEKELLADEKERAEHLMLVDLGRNDLGRVCRPGTVRVDDYSHIER
TH_2725|M.thermoresistible__bu EEDVLLEKELLADEKERAEHLMLVDLGRNDLGRVCRPGTVRVEDYSHIER
Mb1635|M.bovis_AF2122/97 DEDVLLEKELLADDKERAEHLMLVDLGRNDLGRVCTPGTVRVEDYSHIER
Rv1609|M.tuberculosis_H37Rv DEDVLLEKELLADDKERAEHLMLVDLGRNDLGRVCTPGTVRVEDYSHIER
MMAR_2411|M.marinum_M EEDQLLEKELLADQKELAEHLMLVDLGRNDLGRVCIPGTVRVEDYSHIER
MUL_1589|M.ulcerans_Agy99 EEDQLLEKELLADQKELAEHLMLVDLGRNDLGRVCIPGTVRVEDYSHIER
MLBr_01269|M.leprae_Br4923 EEDQLLEKELLADEKERAEHLMLVDLGRNDLGRVCTPGTVRVEDYSHVER
MAV_3177|M.avium_104 EEDQLLEKELLADEKERAEHLMLVDLGRNDLGRVCAPGTVRVDDYSHIER
MAB_2647c|M.abscessus_ATCC_199 EEDVLLAKDLLADEKELSEHLMLVDLGRNDLGRVCTPGSVKVSDYSHIER
:** ** *:**.*:** :***************** **:*:*.****:**
Mflv_3602|M.gilvum_PYR-GCK YSHVMHLVSTVTGLLADGRSALDAVTACFPAGTLSGAPKVRAMELIEEVE
Mvan_2814|M.vanbaalenii_PYR-1 YSHVMHLVSTVTGLLADGKTALDAVTACFPAGTLSGAPKVRAMELIEEVE
MSMEG_3217|M.smegmatis_MC2_155 YSHVMHLVSTVTGELAEDKTALDAVTACFPAGTLSGAPKVRAMELIEEVE
TH_2725|M.thermoresistible__bu YSHVMHLVSTVTGELAEGKTALDPITACFPAGTLSGAPKVRAMELIEEVE
Mb1635|M.bovis_AF2122/97 YSHVMHLVSTVTGKLGEGRTALDAVTACFPAGTLSGAPKVRAMELIEEVE
Rv1609|M.tuberculosis_H37Rv YSHVMHLVSTVTGKLGEGRTALDAVTACFPAGTLSGAPKVRAMELIEEVE
MMAR_2411|M.marinum_M YSHVMHLVSTVTGLLDAGHSALDAVTACFPAGTLSGAPKVRAMELIEEME
MUL_1589|M.ulcerans_Agy99 YSHVMHLVSTVTGLLDAGHSALDAVTACFPAGTLSGAPKVRAMELIEEME
MLBr_01269|M.leprae_Br4923 YSHVMHMVSTVTGLLGEGRTALDAVTACFPAGTLSGAPKVRSMELIEEVE
MAV_3177|M.avium_104 YSHVMHLVSTVTGVLDAGRTALDAVTACFPAGTLSGAPKVRAMELIEEVE
MAB_2647c|M.abscessus_ATCC_199 YSHVMHLVSTVSGSLADGKTALDAITACFPAGTLSGAPKVRAMELIEEVE
******:****:* * .::***.:****************:******:*
Mflv_3602|M.gilvum_PYR-GCK KTRRGLYGGVLGYLDFAGNADFAIAIRTALMRDGTAYVQAGGGVVADSNG
Mvan_2814|M.vanbaalenii_PYR-1 KTRRGLYGGVLGYLDFAGNADFAIAIRTALIRNGTAYVQAGGGVVADSNG
MSMEG_3217|M.smegmatis_MC2_155 KTRRGLYGGVVGYLDFAGNADFAIAIRTALMRNGTAYVQAGGGVVADSNG
TH_2725|M.thermoresistible__bu KTRRGLYGGVLGYLDFAGNADFAIAIRTALIRNGTAYVQSGGGVVADSNG
Mb1635|M.bovis_AF2122/97 KTRRGLYGGVVGYLDFAGNADFAIAIRTALMRNGTAYVQAGGGVVADSNG
Rv1609|M.tuberculosis_H37Rv KTRRGLYGGVVGYLDFAGNADFAIAIRTALMRNGTAYVQAGGGVVADSNG
MMAR_2411|M.marinum_M KTRRGLYGGVVGYLDFAGNADFAIAIRTALMRNGTAYVQAGGGVVADSNG
MUL_1589|M.ulcerans_Agy99 KTRRGLYGGVVGYLDFAGNADFAIAIRTALMRNGTAYVQAGGGVVADSNG
MLBr_01269|M.leprae_Br4923 KTRRGLYGGVVGYLDFAGNADFAIAIRTALMRDGIAYVQAGGGVVADSNG
MAV_3177|M.avium_104 KTRRGLYGGVVGYLDFAGNADFAIAIRTALMRGGTAYVQSGGGVVADSNG
MAB_2647c|M.abscessus_ATCC_199 LTRRGLYGGVLGYLDFAGNADFAIAIRTALFKDGRAYVQAGGGVVADSTG
*********:*******************::.* ****:********.*
Mflv_3602|M.gilvum_PYR-GCK PYEYNESANKARAVLAAIAAAETLSEP-----------------------
Mvan_2814|M.vanbaalenii_PYR-1 PYEYNEASNKARAVLAAIAAAETLSEP-----------------------
MSMEG_3217|M.smegmatis_MC2_155 PYEYTEAANKARAVLNAIAAAATLAEP-----------------------
TH_2725|M.thermoresistible__bu PYEYNEAANKARAVLHAIAAAETLSEP-----------------------
Mb1635|M.bovis_AF2122/97 SYEYNEARNKARAVLNAIAAAETLAAPG----------ANRSGC------
Rv1609|M.tuberculosis_H37Rv SYEYNEARNKARAVLNAIAAAETLAAPG----------ANRSGC------
MMAR_2411|M.marinum_M PYEYNEARNKARAVLNAIAAAETLARPD----------AACSD-------
MUL_1589|M.ulcerans_Agy99 PYEYNEARNKARAVLNAIAAAETLARPD----------AACSD-------
MLBr_01269|M.leprae_Br4923 PYEYIEASNKARAVLNAIAAAETLTSLDFGVALAPGRVAARGEAGNQGRL
MAV_3177|M.avium_104 PYEYTEATNKARAVLGAIAAAETLTEPG----------ARR---------
MAB_2647c|M.abscessus_ATCC_199 EYEYNEARNKATAVLNAIAAAETLTGAADV--------------------
*** *: *** *** ***** **: