For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGRHSAPDPDDFLDEPSPDHPVDERDDAYAFDAQGAPDEGYYPDERRYPDADFVADDDYTPEEFAPGEDL VDEDPDDYPEFPSRRPAPSGSQESPASAPSLRARRLDWRGGHRSEGGRRGVSIGVIVALVAVVVVVGSVI LWRFFGDALSKRSHTAAGRCVGGQEQVPVVADPSIADAIGQFAESFNKSAGPIGDHCMVVSVKPAGSDAV LNGFIGKWPAELGGQPALWIPGSSVSAARLAGATAQKTITESHSLASSPVVLAVRPELLPALSGQNWAAL PGLQTNPNALAGLNLPAWGSLRLALPMTGNGDAAFLAGEAVAAASVPPGAPVTQGTGAVRTLLSAQPKLA DNSLTEAMNTLLKPGDPASAPVHGVVTTEQQLFQRGQSLPDAKGALASWLPPGAAAVADYPTVLLSGSWL TREQASAASEFSRFMHKSDQLAKLAKAGFRVNGGKPPSSPVTTFPALPSTLSVGDDAMRATLAEAMASPS TGQATTIMLDQSMPGQEGGKSRLANVIGALQDKIKALPASAVVGLWTFDGHEGRSEVTSGPLADPVNGQP RSAALSAALDKQYSSSGGAVSFTTLRMIYQDMQSNYHAGQTNSILVITAGPHTDQTLDGPGLQDFIRKSA DPAKPIAVNIIDFGADPDRTTWEAVAQLSGGSYQNLATSASPDLATAVNAFLS
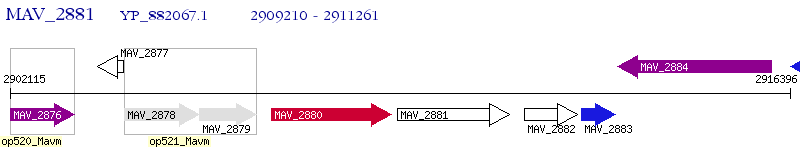
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2881 | - | - | 100% (683) | hypothetical protein MAV_2881 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1867c | - | 0.0 | 68.83% (693) | hypothetical protein Mb1867c |
| M. gilvum PYR-GCK | Mflv_3368 | - | 0.0 | 51.24% (683) | von Willebrand factor, type A |
| M. tuberculosis H37Rv | Rv1836c | - | 0.0 | 68.83% (693) | hypothetical protein Rv1836c |
| M. leprae Br4923 | MLBr_02070 | - | 0.0 | 67.09% (711) | hypothetical protein MLBr_02070 |
| M. abscessus ATCC 19977 | MAB_2408c | - | 1e-152 | 48.18% (577) | hypothetical protein MAB_2408c |
| M. marinum M | MMAR_2712 | - | 0.0 | 60.95% (740) | hypothetical protein MMAR_2712 |
| M. smegmatis MC2 155 | MSMEG_3641 | - | 0.0 | 50.26% (762) | hypothetical protein MSMEG_3641 |
| M. thermoresistible (build 8) | TH_0289 | - | 0.0 | 50.64% (703) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3054 | - | 0.0 | 60.54% (740) | hypothetical protein MUL_3054 |
| M. vanbaalenii PYR-1 | Mvan_3098 | - | 0.0 | 53.95% (684) | von Willebrand factor, type A |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2881|M.avium_104 -----------------------MGRHSAPDPDDFLDEPS-------PDH
MLBr_02070|M.leprae_Br4923 MTKLGAEFKPDQTRINRKAANTGMGRHSMPDPEDSIDQPSNQFAASGPDQ
Mb1867c|M.bovis_AF2122/97 -----------------------MGRHSKPDPEDSVDDLSDGHAAEQQHW
Rv1836c|M.tuberculosis_H37Rv -----------------------MGRHSKPDPEDSVDDLSDGHAAEQQHW
MMAR_2712|M.marinum_M -----------------------MGRHSLPDPEDSADEPSDDHDAENQDW
MUL_3054|M.ulcerans_Agy99 -----------------------MGRHSLPDPEDSADEPSDDHDAENQDW
Mflv_3368|M.gilvum_PYR-GCK -----------------------MGRHSLPDPDEAG------------GR
Mvan_3098|M.vanbaalenii_PYR-1 -----------------------MGRHSLPDPDESD------------QS
MSMEG_3641|M.smegmatis_MC2_155 -----------------------MGRHSIPDPEDAADEPESGYRQGADGQ
TH_0289|M.thermoresistible__bu -----------------------MGRHRLPDPDDEPTGDDSATERFRFPF
MAB_2408c|M.abscessus_ATCC_199 -----------------------MGRHSASGSGSPND-------------
**** ... .
MAV_2881|M.avium_104 P--VDE-------------------------RDDAYAFDAQGAPDEGYYP
MLBr_02070|M.leprae_Br4923 SDEIDHG-----------------YQSRMGYPEPVFEPAATGSPSYRSYP
Mb1867c|M.bovis_AF2122/97 EDIS----------------------------------------GSYDYP
Rv1836c|M.tuberculosis_H37Rv EDIS----------------------------------------GSYDYP
MMAR_2712|M.marinum_M DDELTGQP-------------GGGADSAAADPGAFAHPQTADSAGGYPYP
MUL_3054|M.ulcerans_Agy99 DDELTGQP-------------GGGADSAAADPGAFAHPQTADSAGGYPYP
Mflv_3368|M.gilvum_PYR-GCK RSEP------------------------------------PVPRDGSESD
Mvan_3098|M.vanbaalenii_PYR-1 GSPA------------------------------------RGFGDFGES-
MSMEG_3641|M.smegmatis_MC2_155 GGPPDHPTEVFERIGDSGPATPRRRHRYADEPDEPDEIADSDYTDYSDSD
TH_0289|M.thermoresistible__bu SGRPEE--------------RPGDRGGRHFSPDDFEPDFPPDSPGYGDAP
MAB_2408c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2881|M.avium_104 DERRYP-------------------------------DADFVADDDYTPE
MLBr_02070|M.leprae_Br4923 HGAEHPA------------------------------DSTPEALDETIDY
Mb1867c|M.bovis_AF2122/97 -GVDQP-------------------------------DDGPLSSEGHYSA
Rv1836c|M.tuberculosis_H37Rv -GVDQP-------------------------------DDGPLSSEGHYSA
MMAR_2712|M.marinum_M -GWEQSGDTVGHFGDQEA-------------------DEDSADEDGYWAD
MUL_3054|M.ulcerans_Agy99 -GWEQPGDTVGHFGDQEA-------------------DEDSADEDGYWAD
Mflv_3368|M.gilvum_PYR-GCK ------------------------------------------------SP
Mvan_3098|M.vanbaalenii_PYR-1 ------------------------------------------------AD
MSMEG_3641|M.smegmatis_MC2_155 YVDEYADEYGTEYDPEYGDEYGAEYRGGYRGGFGSEYDTDYSDDYSDDSP
TH_0289|M.thermoresistible__bu GDGGYADTGYGGAEPGYGGEVSP--------------GRGFRDWEADQPD
MAB_2408c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2881|M.avium_104 EFAPGED-------------LVDEDPDDYPEFPSRRPAPSGSQESPASAP
MLBr_02070|M.leprae_Br4923 QSYWAEDR--------NEDLFVDGAADDHPDFPPRPAGSSTSSQAPTSLS
Mb1867c|M.bovis_AF2122/97 VGGYSAS-----------------GSEDYPDIPPRPD-WEPTGAEPIAAA
Rv1836c|M.tuberculosis_H37Rv VGGYSAS-----------------GSEDYPDIPPRPD-WEPTGAEPIAAA
MMAR_2712|M.marinum_M EQVFDESQYLEQDPYGADDRHPELGAEEYPDFGTHPDGPEPSDPKPAATP
MUL_3054|M.ulcerans_Agy99 EQVFDESQYLEQDPYGADDRHPELGAEEYPDFGTHPDGPEPSDPKPAATP
Mflv_3368|M.gilvum_PYR-GCK TDVFG----------------------------DDEQTSRLATGG-----
Mvan_3098|M.vanbaalenii_PYR-1 SGEFGGFRAS-----------------------DTPGSPTAPRSG-----
MSMEG_3641|M.smegmatis_MC2_155 RDEYGYSTASG-------HAPRRRGRDDLSVDDEDAETTKLSAAGGTPPP
TH_0289|M.thermoresistible__bu FGEYGDEPAYG-------DDSYGDDSYGDDAFGDDSYGEDSSDEEPAETP
MAB_2408c|M.abscessus_ATCC_199 --------------------------------PEDNDGWEADSAPGSE--
MAV_2881|M.avium_104 S---LRARRLD----WRGGHRSEG-GRRGVSIGVIVALVAVVVVVGSVIL
MLBr_02070|M.leprae_Br4923 H---LFKASHRSVGKWQGGHRSDG-GRRGVSIGVIATLVAVVVLVGAVIM
Mb1867c|M.bovis_AF2122/97 PPPLFRFG-HRGPGDWQAGHRSAD-GRRGVSIGVIVALVAVVVMVAGVIL
Rv1836c|M.tuberculosis_H37Rv PPPLFRFG-HRGPGDWQAGHRSAD-GRRGVSIGVIVALVAVVVMVAGVIL
MMAR_2712|M.marinum_M PPPLFRVAGHRGLRGWQGGHRSAD-GRRGVSVGVIVALVAVVVVVVGVIG
MUL_3054|M.ulcerans_Agy99 PPSLFRVAGHRGLRGWQGGHRSAD-GRRGVSVGVIVALVAVVVVVVGVIG
Mflv_3368|M.gilvum_PYR-GCK ---GQRSGGWEN-GEWTGSHRAVTPGPRKVSVGVIVALVSVVVVVAAVIL
Mvan_3098|M.vanbaalenii_PYR-1 ---PQHSGGWEG-GEWTGSHRAVTPGRRKVSLGVIVALVAVVVVVATVIV
MSMEG_3641|M.smegmatis_MC2_155 PPRSRRSRSADGDSEWTGSHRAVESRRRGVSVGVIVALVSVVVLVAAVIV
TH_0289|M.thermoresistible__bu TAGRRRVFDPDT-GEWTGSHRAVDSRPRGVSPGVIAALVGVVILVAGFIG
MAB_2408c|M.abscessus_ATCC_199 ----SGSGSEFDTGSWQRSHRSGG-SNWGVSKGLIGAVAAVLVVAVSIGL
* .**: ** *:* ::..*:::. .
MAV_2881|M.avium_104 WRFFGDALSKRSHTAAGRCVGGQEQVPVVADPSIADAIGQFAESFNKSAG
MLBr_02070|M.leprae_Br4923 WSFLGHILNNRKHQAAARCVGGHQTVAVVADPSIADYLQEFAQSYNASAR
Mb1867c|M.bovis_AF2122/97 WCFFGDALSNRSHTAAARCVGGKDTVAVIADPSIADQVKESADSYNASAG
Rv1836c|M.tuberculosis_H37Rv WRFFGDALSNRSHTAAARCVGGKDTVAVIADPSIADQVKESADSYNASAG
MMAR_2712|M.marinum_M WRFFGDVLSNRSQTAAARCVGGNDTVAVIADPSIADQVNDFADSYNASSG
MUL_3054|M.ulcerans_Agy99 WRFFGDVLSNRSQTAAARCVGGNDTVAVIADPSIADQVNDFADSYNASSG
Mflv_3368|M.gilvum_PYR-GCK WRFVGDTLSDRSDIAAARCVEGEVAVAVIADPAIADPVAALAQRYNETAD
Mvan_3098|M.vanbaalenii_PYR-1 WRFVGDALSGRSDVAAARCVEGEVAVAVVADPAIAEPVAALAERYNETAA
MSMEG_3641|M.smegmatis_MC2_155 WKFVGDALSDRSDAAAARCVAGEIGVPVIADPTISTHIESLANKYNQSAS
TH_0289|M.thermoresistible__bu WQFFGDALSNRSGAAAARCVAGDLNISVLADPSISDHTRTLADRYNDTAS
MAB_2408c|M.abscessus_ATCC_199 WWYFDRRTSDNQAEAAATCVHGNNAIAVVADPSIADRIGELSERFNQKHE
* :.. . .. **. ** *. :.*:***:*: :: :* .
MAV_2881|M.avium_104 PIGDHCMVVSVKPAGSDAVLNGFIGKWPAELGGQPALWIPGSSVSAARLA
MLBr_02070|M.leprae_Br4923 PVGDHCMMVTVKPVGSEAALTGFNDSWPANLGDKPALWIPGSSISAARLA
Mb1867c|M.bovis_AF2122/97 PVGDRCVAVAVTSAGSDAVINGFIGKWPTELGGQPGLWIPSSSISAARLT
Rv1836c|M.tuberculosis_H37Rv PVGDRCVAVAVTSAGSDAVINGFIGKWPTELGGQPGLWIPSSSISAARLT
MMAR_2712|M.marinum_M PIGDRCVSVAVKAADADAVITGFIGKWPSELGAQPGLWIPGSSVSAARLV
MUL_3054|M.ulcerans_Agy99 PIGDRCVSVAVNAADADAVITGFIGKWPSELGAQPGLWIPGSSVSAARLV
Mflv_3368|M.gilvum_PYR-GCK PVGDRCVKVGVTPADSGRVVNGFGEQWPGDLGERPALWIPASSVSEARLE
Mvan_3098|M.vanbaalenii_PYR-1 PVGDRCVKVGVKSADSDQVLNGFSGQWPGDLGERPALWIPASSVSGARLE
MSMEG_3641|M.smegmatis_MC2_155 PVGDKCVKVRVQSAESDRVVSGFANSWPSELGDRPALWIPSSSIGSARLE
TH_0289|M.thermoresistible__bu QVGDKCVKVSVRSAEPDAVIDGLGSQWPADLGDRPAVWIPASSLSAARLE
MAB_2408c|M.abscessus_ATCC_199 VIGDYCFTVSVRPADSANVIKGLTGQWPAELGEQPALWIPGSSISSARLK
:** *. * * .. . .: *: .** :** :*.:***.**:. ***
MAV_2881|M.avium_104 GATAQKTITESHSLASSPVVLAVRPELLPALSG-QNWAALPGLQTNPNAL
MLBr_02070|M.leprae_Br4923 VTADQKTISESHSLVTSPVLLAVRPEFEQALAN-KGWAALPGLQTNPNSL
Mb1867c|M.bovis_AF2122/97 GAAGSQAISDSRSLVISPVLLAVRPELQQALAN-QNWAALPGLQTNPNSL
Rv1836c|M.tuberculosis_H37Rv GAAGSQAISDSRSLVISPVLLAVRPELQQALAN-QNWAALPGLQTNPNSL
MMAR_2712|M.marinum_M QAAGKEAISDSRSLVTSPVLLAIRPELQQALGN-QNWAALPGLQSDPNSM
MUL_3054|M.ulcerans_Agy99 QAAGKEAISDSRSLVTSPVLLAIRPELQQALGN-QNWAAVPGLQSDPNSM
Mflv_3368|M.gilvum_PYR-GCK ASTGPETISDSRSLVTSPVLLAVAAELKDALGE-RDWGSLPDLQSNPNSL
Mvan_3098|M.vanbaalenii_PYR-1 AATGAETVSDSRSLVTSPVVLAVAPALKDALGQ-QNWGTLPRLQTDPAAL
MSMEG_3641|M.smegmatis_MC2_155 ATAGSETVSDSRSLVTSPVVLATSPELKTALGQ-QNWQKLPELQSSPTAM
TH_0289|M.thermoresistible__bu AAAGPQTIDDSRSLVTSPVLLAVRPELKFALGQ-RDWSALPDLQRNPTAL
MAB_2408c|M.abscessus_ATCC_199 AASKTNIVSDSRSLVSTPVVIAVTPKLRQAIPNDKSWADVPALQNVPNSL
:: : : :*:**. :**::* . : *: :.* :* ** * ::
MAV_2881|M.avium_104 AGLNLPAWGSLRLALPMTGNGDAAFLAGEAVAAASVPPGAPVTQGTGAVR
MLBr_02070|M.leprae_Br4923 ADLNLPAWGSLRLALPMNGNSDATFLAGEAVATASVPAGAPAIQGVGAVR
Mb1867c|M.bovis_AF2122/97 SGLDLPAWGSLRLAMPSSGNGDAAYLAGEAVAAASAPAGAPATAGIGAVR
Rv1836c|M.tuberculosis_H37Rv SGLDLPAWGSLRLAMPSSGNGDAAYLAGEAVAAASAPAGAPATAGIGAVR
MMAR_2712|M.marinum_M AGLKLPSWGSLRLALPVGGNGDATFLAGEAVAAASAPADAPPTAGIGAVR
MUL_3054|M.ulcerans_Agy99 AGLKLPSWGSLRLALPVGGNGDATFLAGEAVAAASAPADAPPTAGIGAVR
Mflv_3368|M.gilvum_PYR-GCK DGLGLRGWGSLRLAMPLGDDSDASFLAAEAVAAAAAPAGEPATAGLGAVS
Mvan_3098|M.vanbaalenii_PYR-1 DGLGLQGWGGLRLALPLGDDSDASYLAAEAIAAAAAPSGAPASAGLGAVS
MSMEG_3641|M.smegmatis_MC2_155 DGLRLPNWGTLKLALPKLDNGDTAYLVAEAVAVTSAPSGAPPTAGMGAVS
TH_0289|M.thermoresistible__bu DDLGLNGWGSLKLALPRSGGADASGLVAESVAAASAPDGAPATGGLGAVN
MAB_2408c|M.abscessus_ATCC_199 DGVGLPGWGSLRLALPSSGNADAAQLAAEAVAAASVRPGDSPELGAGAAG
.: * ** *:**:* ...*:: *..*::*.::. . . * **.
MAV_2881|M.avium_104 TLLSAQPKLADNSLTEAMNTLLKPGDPASAPVHGVVTTEQQLFQRGQSLP
MLBr_02070|M.leprae_Br4923 TLMSAQPKLADSTWAEAMSTLLKPGDVATAPVHAVITTEQQLFQRGQSLS
Mb1867c|M.bovis_AF2122/97 TLMGARPKLADDSLTAAMDTLLKPGDVATAPVHAVVTTEQQLFQRGQSLS
Rv1836c|M.tuberculosis_H37Rv TLMGARPKLADDSLTAAMDTLLKPGDVATAPVHAVVTTEQQLFQRGQSLS
MMAR_2712|M.marinum_M TLMATQPKLADGSLSEAMDALLKADDVAAAPVHAVITTEQQLFLRAQSLS
MUL_3054|M.ulcerans_Agy99 TLMATQPKLADGSLSEAMDALLKADDVAAAPVHAVITTEQQLFLRAQSLS
Mflv_3368|M.gilvum_PYR-GCK TLLSRAPELSDADAGTALDALADASDNAAAPVHAVVTTEQRVFQRASTAP
Mvan_3098|M.vanbaalenii_PYR-1 TVMSGAPELADPNAGTAIDALVGAADQAAAPVHAVVTTEQRVFQRASSLP
MSMEG_3641|M.smegmatis_MC2_155 TLLNGQPKLDDAELSTAFDAMLDPSDSAAAPVHAVATTEQQLFQRATTLD
TH_0289|M.thermoresistible__bu ALLAGQPELSDDTWSTAFDALVDSGDPRSAPVHAVASTEQQLYQRSTSLS
MAB_2408c|M.abscessus_ATCC_199 SLAATAPKLPANNVADAIGALLDGGEQPGAAVHAVVTTEQQLYARTRNNG
:: *:* *:.:: : *.**.* :***::: * .
MAV_2881|M.avium_104 DAKGALASWLPPGAAAVADYPTVLLSGSWLTREQASAASEFSRFMHKSDQ
MLBr_02070|M.leprae_Br4923 DAKSALGSWLPHGPAPVADYPAVLLNGSWLTQEQAAAASEFARFVQKPDQ
Mb1867c|M.bovis_AF2122/97 DAENTLGSWLPPGPAAVADYPTVLLSGAWLSQEQTSAASAFARYLHKPEQ
Rv1836c|M.tuberculosis_H37Rv DAENTLGSWLPPGPAAVADYPTVLLSGAWLSQEQTSAASAFARYLHKPEQ
MMAR_2712|M.marinum_M DAKKKLSSWLPPGPVAVADYPAVLLNGSWLSQEQTTAASAFARYVHKPEQ
MUL_3054|M.ulcerans_Agy99 DAKKKLSSWLPPGPVAVADYPAVLLNGSWLSQEQTTAASAFARYVHKPEQ
Mflv_3368|M.gilvum_PYR-GCK DADSKPAAWLPSGPAVLADFPTVLLSGDWLSQEQVTGASEFARFLRKPEQ
Mvan_3098|M.vanbaalenii_PYR-1 DSKDKLAAWIPPGPTATADFPTVLLAGDWLSQEQVTAASEFARFMRKPEQ
MSMEG_3641|M.smegmatis_MC2_155 DAGSKLAGWLPQGPAAVADYPTVLLAGSWLEQEQVTAASEFARYLRKPEQ
TH_0289|M.thermoresistible__bu DPAQTVAAFRPDGPTAVADYPTVLLSGDWLEQDQRAAASEFVRFLRRPEQ
MAB_2408c|M.abscessus_ATCC_199 DAKKVIAQWQPAGATPIADYPTVQLDGAWLSEEQHTAASQFARFLGDKDQ
*. . : * *.. **:*:* * * ** .:* :.** * *:: :*
MAV_2881|M.avium_104 LAKLAKAGFRVNGGKPPSSPVTTFPALPSTLSVGDDAMRATLAEAMASPS
MLBr_02070|M.leprae_Br4923 LAKLAKAGFRVNGVTPPSSSVTSFAAVPSTVSVGDDGMRATLVEEMIQPS
Mb1867c|M.bovis_AF2122/97 LAKLARAGFRVSDVKPPSSPVTSFPALPSTLSVGDDSMRATLADTMVTAS
Rv1836c|M.tuberculosis_H37Rv LAKLARAGFRVSDVKPPSSPVTSFPALPSTLSVGDDSMRATLADTMVTAS
MMAR_2712|M.marinum_M LAKLAKAGFRVDDVKPPSSPVTSFPALPAPLSVGDEGIRATLADAVAAPS
MUL_3054|M.ulcerans_Agy99 LAKLAKAGFRVDDVKPPSSPVTSFPALPAPLSVGDEGIRATLADAVAAPS
Mflv_3368|M.gilvum_PYR-GCK LGELAKAGFRVEGVEPPASDVVDFAPLSAPLAVGDNQVRTTIADTLTMPV
Mvan_3098|M.vanbaalenii_PYR-1 LGELAKAGFRVEGTAPPASDVVDFAPVSAPLEVGDNALRSTIAETLATPV
MSMEG_3641|M.smegmatis_MC2_155 LAELAKAGFRAEDATSPDSDVTDFGPIANPVSIADESTRVTLANATAAPV
TH_0289|M.thermoresistible__bu LGELAAAGFRTDAAGPPDSAVVDFAAVSTPLSLEDDEIRVTLAEAVTSPQ
MAB_2408c|M.abscessus_ATCC_199 IKDLAAAGFRAEGTDLPTSDVVSFAKIDKPLSIEDK-VRVALADGTSTGS
: .** ****.. * * *. * : .: : *. * ::.:
MAV_2881|M.avium_104 TGQATTIMLDQSMPGQEGGKSRLANVIGALQDKIKALPASAVVGLWTFDG
MLBr_02070|M.leprae_Br4923 SGVAATIMLDQSMPTDEGGKTRLANVVAALDDKINAMPPTSVMGLWTFDG
Mb1867c|M.bovis_AF2122/97 AGVAATIMLDQSMPNDEGGNSRLSNVVAALENRIKAMPPSSVVGLWTFDG
Rv1836c|M.tuberculosis_H37Rv AGVAATIMLDQSMPNDEGGNSRLSNVVAALENRIKAMPPSSVVGLWTFDG
MMAR_2712|M.marinum_M MGVAATIMLDQSLSTDDGGKTRLTNIVAALQNRIKTLPPTSAVGLWTFDG
MUL_3054|M.ulcerans_Agy99 MGVAATIMLDQSLSTDDGGKTRLTNIVAALQNRVKTLLPTSAVGLWTFDG
Mflv_3368|M.gilvum_PYR-GCK ETSTVTVMLDQSMPVDEGGATRLANVVDALKARIPVLPPDSGVGLWTFDG
Mvan_3098|M.vanbaalenii_PYR-1 GSPTVTVMLDQSMPVEEGGVSRLQNVIDALKARIAVLPADSGVGLWTFDG
MSMEG_3641|M.smegmatis_MC2_155 SSPAVTIMLDRSMPTDEGGRSRLQNVVEALTNRLKALPVTSEVGLWTFDG
TH_0289|M.thermoresistible__bu AGAAVTIMLDRSMTEVEGSNTRMGNAVNALIDRVPALPPSAAVGLWTFDG
MAB_2408c|M.abscessus_ATCC_199 G--TTTIMLASSPAPD----AKLSDITGPLANRVRALAPGSGIGLWVYDG
:.*:** * . ::: : .* :: .: : :***.:**
MAV_2881|M.avium_104 HEGRSEVTSGPLADPVNGQPRSAALSAALDKQYSSSGGAVSFTTLRMIYQ
MLBr_02070|M.leprae_Br4923 HKGQTEVTTGQLADPVNGQPRSAALTAALDKQYSSNGGAVSFTTLRMIYQ
Mb1867c|M.bovis_AF2122/97 REGRTEVPAGPLADPVNGQPRPAALTAALGKQYSSGGGAVSFTTLRLIYQ
Rv1836c|M.tuberculosis_H37Rv REGRTEVPAGPLADPVNGQPRPAALTAALGKQYSSGGGAVSFTTLRLIYQ
MMAR_2712|M.marinum_M REGRTEVPTGPLADPVNGQPRSAALNAALGKQYSSNGGAVSFTTLRMIYE
MUL_3054|M.ulcerans_Agy99 REGRTEVPTGPLADPVNGQPRSAALNAALDKQYSSNGGAVSFTTLRMIYE
Mflv_3368|M.gilvum_PYR-GCK VAGRSEVAVGPLADPVDGTPRSEVLTAALDRQTASGGGAVSFTTLRLVYG
Mvan_3098|M.vanbaalenii_PYR-1 VQGRSAVSVGPLSEPVDGAPRKEALTAALDSQSPSGGGAVSFTTLRLVYT
MSMEG_3641|M.smegmatis_MC2_155 TEGRSEVSMGPMAEPVDGRARSEVLNSTLEDQSAAGGGAVSFTTLRLVYN
TH_0289|M.thermoresistible__bu VAGRSEMTVGPLSEPVDGRPRADALTSVLDAQSSTAGGTVSFTTLRLVYT
MAB_2408c|M.abscessus_ATCC_199 KEGNTVVRLGGAGDDVEGMPRSQSVADALTALQPTGNGAVAYTTLRALYQ
*.: : * .: *:* .* : .* .: .*:*::**** :*
MAV_2881|M.avium_104 DMQSNYHAGQTNSILVITAGPHTDQTLDGPGLQDFIRKSADPAKPIAVNI
MLBr_02070|M.leprae_Br4923 EMLANYHVGQTNSVLVITAGPHTDQTLDGARLQDFIRTSADPAKPIAVNV
Mb1867c|M.bovis_AF2122/97 EMLANYHVGQANSVLVITAGPHTDQTLDGPGLQDFIRKSADPAKPIAVNI
Rv1836c|M.tuberculosis_H37Rv EMLANYRVGQANSVLVITAGPHTDQTLDGPGLQDFIRKSADPAKPIAVNI
MMAR_2712|M.marinum_M DVQAHFRADQANSILVITGGPHTDQSLDGPGLENFIRTSADPAKPIAVNV
MUL_3054|M.ulcerans_Agy99 DVQAHFRADQANSILVITGGPHTDQSLDGPGLENFIRTSADPAKPIAVNV
Mflv_3368|M.gilvum_PYR-GCK DATARFREGQKNSVLVITTGPHTDRSLGAQGLQDYIRGAFTPDRPVAVNV
Mvan_3098|M.vanbaalenii_PYR-1 DASTKYREGQKNSVLVITTGPHTDQSLGAAGLQDYIRGAFNRDRPVAVNV
MSMEG_3641|M.smegmatis_MC2_155 EAKANFVEGRGNSVLVITTGPHTDRSLDGPGLQEFIRSNFDPARPIAVNV
TH_0289|M.thermoresistible__bu DATANYQDGQPNSVLVITGGPHTDRTLDGPGLQQFLRQTFDPQRPIAVNI
MAB_2408c|M.abscessus_ATCC_199 DAVAGFRPNQVNSVLVVAGRSHTDQTLDGPGLIDTINRLKDPAKPVRINV
: : : .: **:**:: .***::*.. * : :. :*: :*:
MAV_2881|M.avium_104 IDFGADPDRTTWEAVAQLSGGSYQNLATSASPDLATAVNAFLS
MLBr_02070|M.leprae_Br4923 IDFGTDPDQATWKAVAQISGGSYQNLSTSASLDLATAINTFLS
Mb1867c|M.bovis_AF2122/97 IDFGADPDRATWEAVAQLSGGSYQNLETSASPDLATAVNIFLS
Rv1836c|M.tuberculosis_H37Rv IDFGADPDRATWEAVAQLSGGSYQNLETSASPDLATAVNIFLS
MMAR_2712|M.marinum_M IDFGADPDRKTWEAVAQLSGGSYQNLATSTGPNLAAAVDTFLS
MUL_3054|M.ulcerans_Agy99 IDFGADPDRKTWEAVAQLSGGSYQNLATSTGPNLAAAVDTFLS
Mflv_3368|M.gilvum_PYR-GCK IDFGDDADRPTWESVAEITGGSYRNMADSTSPDLSSAISEMLG
Mvan_3098|M.vanbaalenii_PYR-1 IDFGDDSDRATWESVAQITGGNYQNLGTSASPELAAAISSMLS
MSMEG_3641|M.smegmatis_MC2_155 IDFGDDSDRETWEAVAQASGGDYVNLPTSTAPELVTSVATMLG
TH_0289|M.thermoresistible__bu IDFGEDSDVPTWQAVAEATGGTYVNLPTSDTPELAATITRLFG
MAB_2408c|M.abscessus_ATCC_199 LDFGADSDQQTWQTIAQQSGGAYQNVSASNSPELAAAIARFIS
:*** *.* **:::*: :** * *: * :* ::: ::.