For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTVRIGTSGWSYDHWADVLYPPGTPSARRLARYIEVFDTVELNASFYRWPKDSTFAGWREQLPDGFTMSV KAHRGLTHYRRLASPEPWIERFERCWELLGDRRGVLLVQLHPEQQRDDARLDSFLERMPASIRVAVELRH PSWNDPAVYALLERRRAAYVVTSGLGLACIPRATTDLVYIRMHGPDPAANYAGSYSDDDLRCWAERITAW DGDGKDVWMYFNNDLGGHAVRNALALRELVG
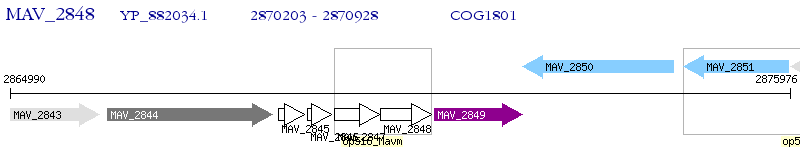
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2848 | - | - | 100% (241) | hypothetical protein MAV_2848 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0717 | - | 2e-93 | 67.09% (237) | hypothetical protein Mflv_0717 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3593 | - | 1e-100 | 71.85% (238) | hypothetical protein MSMEG_3593 |
| M. thermoresistible (build 8) | TH_1433 | - | 8e-98 | 67.90% (243) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0127 | - | 1e-94 | 68.05% (241) | hypothetical protein Mvan_0127 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3593|M.smegmatis_MC2_155 -MIRIGTSGWSYDHWRDVLYPPGTPSSARLSRYTQVFDTVELNASYYRWP
TH_1433|M.thermoresistible__bu -MIRIGTSGWSYDHWTGVLYPPRTPPGRRLAVYAAEFDTVELNASFYRWP
MAV_2848|M.avium_104 MTVRIGTSGWSYDHWADVLYPPGTPSARRLARYIEVFDTVELNASFYRWP
Mflv_0717|M.gilvum_PYR-GCK MSVRIGTSGWAYTHWRGVLYEPGLPTTRWLQRYVDEFDTVELNGSFYRWP
Mvan_0127|M.vanbaalenii_PYR-1 MTVRVGTSGWSYDHWRNVLYEPGLPAARRLERYAAEFDTVELNGSFYRWP
:*:*****:* ** .*** * *. * * *******.*:****
MSMEG_3593|M.smegmatis_MC2_155 RDTAFANWRSQLPDGFTMSVKAHRGLTHFRRLRSPEPWIERFERCWEVLD
TH_1433|M.thermoresistible__bu RDSTFAGWRQRLPDGFTMSVKAHRGLTHFRRLTGPEPWVERFRRCWDALG
MAV_2848|M.avium_104 KDSTFAGWREQLPDGFTMSVKAHRGLTHYRRLASPEPWIERFERCWELLG
Mflv_0717|M.gilvum_PYR-GCK SDAQFERWRDQLPDGFVMAVKAARGLTHARRLRSPEVWVERLERGWRALG
Mvan_0127|M.vanbaalenii_PYR-1 SDAQFQRWRDQLQPGFVMAVKAARGLTHARRLRTPEVWIERLERGWRALG
*: * **.:* **.*:*** ***** *** ** *:**:.* * *.
MSMEG_3593|M.smegmatis_MC2_155 GCAEALLVQPHPALER--DD--ALLDEFLTLVPDHIPVAVELRHASWHDP
TH_1433|M.thermoresistible__bu DRAEALLVQLHPDFERGQHDGLERLDHFLTVMPDHIPVAVEIRHPSWDAP
MAV_2848|M.avium_104 DRRGVLLVQLHPEQQR--DD--ARLDSFLERMPASIRVAVELRHPSWNDP
Mflv_0717|M.gilvum_PYR-GCK ERKGPLLVQLHPALER--DD--ARLDRFLGVMPADIPVAVEFRHPSWDDP
Mvan_0127|M.vanbaalenii_PYR-1 DRGGPLLVQLHPALER--DD--ARLDHFLGAMSAGIPVAVEFRHPSWDDP
**** ** :* .* ** ** :. * ****:**.**. *
MSMEG_3593|M.smegmatis_MC2_155 AVYALLRRHGAAYVVMSGPGLVCEPVATSDLVYIRLHGPPQEAIYSGSYP
TH_1433|M.thermoresistible__bu EVFRLLERHGAGYVVMSGAGLRCIPRATGRLVYIRLHGPDQHSRYHGSYP
MAV_2848|M.avium_104 AVYALLERRRAAYVVTSGLGLACIPRATTDLVYIRMHGPDPAANYAGSYS
Mflv_0717|M.gilvum_PYR-GCK QVYELLERHDAAYVVMSGAHLPCILKATASLVYVRLHGPDPDALYGGSYS
Mvan_0127|M.vanbaalenii_PYR-1 AVYELLERHDAAYVVMSGGGLPCILKATAGFSYVRLHGPDPQAMYGGSYS
*: **.*: *.*** ** * * ** : *:*:*** : * ***.
MSMEG_3593|M.smegmatis_MC2_155 ADQLRWWAEHILLWHRDGHRVVVYFNNDLGGHAVRNARTLKELVAHE---
TH_1433|M.thermoresistible__bu EHELRRWADRIRSWQDEGRDVDVYFNNDLGGYAVRNARTLRRLLGDDRAT
MAV_2848|M.avium_104 DDDLRCWAERITAWDGDGKDVWMYFNNDLGGHAVRNALALRELVG-----
Mflv_0717|M.gilvum_PYR-GCK ADDLRWWADRIREWTAQGRDVVVYFNNDLGGNAVRNARDLKAALSR----
Mvan_0127|M.vanbaalenii_PYR-1 ADDLRWWAARIGEWDAQGRDVYVYFNNDLGGNAVRNARDLKAELGRRPEG
.:** ** :* * :*: * :******** ***** *: :.
MSMEG_3593|M.smegmatis_MC2_155 --------------------------------------------------
TH_1433|M.thermoresistible__bu VVPGEKRVSEVGRPADGGSADRVPDADRLRRSGRGDHRGRRGAAGCGPAG
MAV_2848|M.avium_104 --------------------------------------------------
Mflv_0717|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0127|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3593|M.smegmatis_MC2_155 ----------
TH_1433|M.thermoresistible__bu PGDQHRPPQQ
MAV_2848|M.avium_104 ----------
Mflv_0717|M.gilvum_PYR-GCK ----------
Mvan_0127|M.vanbaalenii_PYR-1 ----------