For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GQTAAMAEADRTWMVDTLLALLQTPSPSGRTDAVMQVIGDILDDFGVPFTLTRRGALTAELPGESATTDR ALVVHSDTIGCMVRGLKDNGRLELVPVGTFSARFAAGARVRIFIDDPEEFITGTILPLKSSGHAFGDEID TQPTGWEHVEVRIDRKVSCRDDLVRLGLQIGDFVALITSPELTADGFVVSRHLDGKAGVAIALALAKNFA ENKVVLPHRTTIMVTITEEVGHGASHGLPPDVAELISVDNAVCAPGQHSLEDGVTIPMADMHGPFDYHLT RKLCRLAQDRGIPFARDIFRYYRSDAAAAIEAGANTRAALVGFGLDGSHGWERTHIDSLEAAYNLLHAWL QTPLTFAKWDAKPTGSLRDFPSSKQPAPSERWVPLSRGDYESPGDASPGTHWPPSEGPQS*
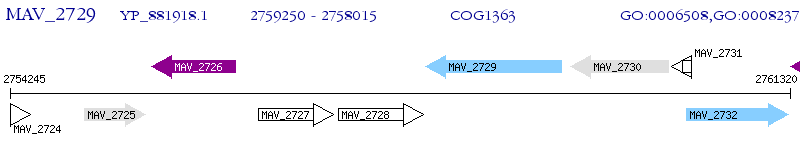
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2729 | - | - | 100% (411) | hydrolase, peptidase M42 family protein |
| M. avium 104 | MAV_1117 | - | 1e-11 | 24.33% (374) | M42 glutamyl aminopeptidase superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3273 | - | 3e-07 | 23.12% (333) | glutamyl aminopeptidase, M42 family protein |
| M. thermoresistible (build 8) | TH_1058 | - | 8e-09 | 22.88% (306) | glutamyl aminopeptidase, M42 family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3273|M.smegmatis_MC2_155 ---------------MADDGFPRA-----------------LLQELLSAY
TH_1058|M.thermoresistible__bu VEYTFGGAFLGVGGAFSGVGARRAGVDGVRAGYVGIMLRADLLQELLVAY
MAV_2729|M.avium_104 ---------MGQTAAMAEADRTWMVD---------------TLLALLQTP
:: . * ** :
MSMEG_3273|M.smegmatis_MC2_155 GPCGQEAAVRVVCRRELEPYVDGLWTDAAGNLVGLLRGAPESADHVPP--
TH_1058|M.thermoresistible__bu GPGGQEDAVRAICHRELAPHVDEIWTDPAGNLVGLIRSADPECPDSPSGP
MAV_2729|M.avium_104 SPSGRTDAVMQVIGDILDDFGVPFTLTRRGALTAELPGESATTD------
.* *: ** : * . : * *.. : .
MSMEG_3273|M.smegmatis_MC2_155 -------TRVMAHMDELAMLVKRVEPDGTLALTPLGTMYPANFGLGPVAM
TH_1058|M.thermoresistible__bu NKDGSQSIRVMAHMDELSMLVKRIDADGSLWLTPAGTMYPGNFGLGPVAV
MAV_2729|M.avium_104 -------RALVVHSDTIGCMVRGLKDNGRLELVPVGTFSARFAAGARVRI
::.* * :. :*: :. :* * *.* **: . . . * :
MSMEG_3273|M.smegmatis_MC2_155 LGERETLCGVLALGSEHTTAESPRIWATKPDQGDKALDWPDVYVFTGRTP
TH_1058|M.thermoresistible__bu LGDQETLTAVLTLGSEHTTKESQRIWETKPDQGDKSLDWLHVYVFTGRTP
MAV_2729|M.avium_104 FIDD---PEEFITGTILPLKSSGHAFGDEIDTQPTGWEHVEVRIDRKVSC
: : : *: . .* : : : * .. : .* : :
MSMEG_3273|M.smegmatis_MC2_155 -DELRDAGIGIGTRVCIDRSKRTLVEVGDYLGCYFMDDRAVVTALLQTAR
TH_1058|M.thermoresistible__bu -EELSAAGVRPGTRVCVHHSKRTLIEFGDYLGCYFLDDRAAVAALLEVAP
MAV_2729|M.avium_104 RDDLVRLGLQIGDFVALITS--PELTADGFVVSRHLDGKAGVAIALALAK
::* *: * *.: * . : ..:: . .:*.:* *: * *
MSMEG_3273|M.smegmatis_MC2_155 LLHGRDHRPPGDTYFVFTTNEEIGGSGGSYASRTLPGDLTLALEVGPTEQ
TH_1058|M.thermoresistible__bu GLR--DPRPPQDVYLVFTTAEEVGGIGGCYASRTLPGRLTLALEVGPTEA
MAV_2729|M.avium_104 NFAENKVVLPHRTTIMVTITEEVGHG----ASHGLPPDVAELISVDNAVC
: . * . ::.* **:* **: ** :: :.*. :
MSMEG_3273|M.smegmatis_MC2_155 EYGTR-VGGGPVVAYSDAQCVYDKDVADRLVQIGEKLGLDPQPAVLGAFE
TH_1058|M.thermoresistible__bu EYGTT-VSGGPIIAYGDAQALYDKKVADRLLAVAIELGLDPQAAVFGAFE
MAV_2729|M.avium_104 APGQHSLEDGVTIPMADMHGPFDYHLTRKLCRLAQDRGIPFARDIFRYYR
* : .* :. .* : :* .:: :* :. . *: :: :.
MSMEG_3273|M.smegmatis_MC2_155 SDASHAKSNGQSPRAGLLCMPTLSTHGYEVIARTAVRAMADVLVEFLADD
TH_1058|M.thermoresistible__bu SDASQARAAGLTAQAALLCLPTLSTHGYEVLARAAIPAMSRVVCEFLRTP
MAV_2729|M.avium_104 SDAAAAIEAGANTRAALVGFGLDGSHGWERTHIDSLEAAYNLLHAWLQTP
***: * * ..:*.*: : .:**:* :: * :: :*
MSMEG_3273|M.smegmatis_MC2_155 RTRR----------------------------------------------
TH_1058|M.thermoresistible__bu QPD-----------------------------------------------
MAV_2729|M.avium_104 LTFAKWDAKPTGSLRDFPSSKQPAPSERWVPLSRGDYESPGDASPGTHWP
.
MSMEG_3273|M.smegmatis_MC2_155 -------
TH_1058|M.thermoresistible__bu -------
MAV_2729|M.avium_104 PSEGPQS