For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSANWGRARAVADAVLYEGYLLYPYRATSSKNESRWQFGVLGPPGAADAGIGEDDSLSAQFLVDHAEAIT LVVRFLHLQHRGAERDTGHGRFEPVGELTAGASSWLTWDEAVETELSFGPLDLTGTQHQWTLPVAAPAAT DIEPLVGGRLVRTREDVSAALSVSVEHDDGLDLVTVAVRNTGAAVDGTSDKDSTIARSLIGTHIIAEAIG GEFISLLETPEHAAAAAARCRQHRCFPVLAGALGEHDLLLVSPIILYDHPEVAEQSEGALYDSTEIDEIL TLRVMTMTDQEKAQARATDPLAAEIIDRCDAMSPEAMQRMHGVLRDPHTGYPASDTGLIPEIPDGLGWWD PEADNAVRPDVDAILINGVRVARGSRVRLHPGRRADAQDLFVDGKSARVTSVHEDVDGNQHVGVVVDDDP DADLHDWYGRYLYFAPDEVEPLEAHPTGTERNPTWR
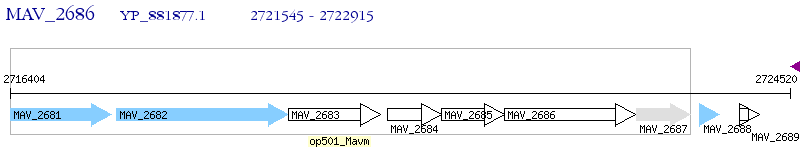
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2686 | - | - | 100% (456) | hypothetical protein MAV_2686 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1865 | - | 0.0 | 70.24% (457) | hypothetical protein MMAR_1865 |
| M. smegmatis MC2 155 | MSMEG_2715 | - | 1e-150 | 60.49% (448) | hypothetical protein MSMEG_2715 |
| M. thermoresistible (build 8) | TH_1949 | - | 1e-168 | 65.52% (464) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2410 | - | 1e-170 | 68.47% (444) | hypothetical protein Mvan_2410 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2686|M.avium_104 MSANWGRARAVADAVLYEGYLLYPYRATSSKNESRWQFGVLGPPGAADAG
MMAR_1865|M.marinum_M MTSNRDRARAVADAVLYEGYLLYPYRGTSSKNQSRWQFGVLGPPGAADAG
Mvan_2410|M.vanbaalenii_PYR-1 MSTGWDRARAVADAVLYEGYLLYPYRATSGKNQSRWQFGVLGPPGAEQSG
TH_1949|M.thermoresistible__bu MSSDEDRVRAVADAVLYEGYLLYPYRADSRKNQTRWQFGVLGPPGAQAAG
MSMEG_2715|M.smegmatis_MC2_155 MSTATEQVLAVADAVLYEGYLLYPYRGDAAKNQCRWQWGVLGPEGAESAG
*:: :. *****************. : **: ***:***** ** :*
MAV_2686|M.avium_104 IGEDDSLSAQFLVD--HAEAITLVVRFLHLQHRGAER----DTGHGRFEP
MMAR_1865|M.marinum_M LGEDDTLAAEFLVE--GTQAITVAVRFLQLQHRRAER----EIN-GAFEA
Mvan_2410|M.vanbaalenii_PYR-1 VGEHDALSAQVLVRPLGEPTVSGVVRFLQLQHRGVER----DLG-DRYEP
TH_1949|M.thermoresistible__bu IGENDSLATQFLVYSGPATTLTLTVRFLQLQHRSAER----ATA-TGYTP
MSMEG_2715|M.smegmatis_MC2_155 IGEDSSMSAQLLVRGDGRARLSVTVRFLQLQHRSAERRHLDDAAHESFVP
:**..:::::.** :: .****:**** .** : .
MAV_2686|M.avium_104 VGELTAGASSWLTWDEAVETELSFGPLDLTGTQHQWTLPVAAPAATDIEP
MMAR_1865|M.marinum_M VDELATPSGSWLTWDEAVECEMSFGPLAFDDPDQPWQLPVLAPAATDIEE
Mvan_2410|M.vanbaalenii_PYR-1 VGELQAGAHRWLRWDEAVECEVAIDAVPVP--ALPRVVEIGVPAGNDVET
TH_1949|M.thermoresistible__bu VRALTAGDRSWLSWDEAVEHELRFGPLPLT--AAPLRHRISVPDGADVES
MSMEG_2715|M.smegmatis_MC2_155 VDELVSTAGSWLSWDEAVAQQVTLGPFDVDDLTTSRTHPIIVEGGREIET
* * : ** ***** :: :... . : . . ::*
MAV_2686|M.avium_104 LVGGRLVRTREDVSAALSVSVEH----DDGLDLVTVAVRNTGAAVDGTSD
MMAR_1865|M.marinum_M VEGGRLVRQRREVRGELTVSSQS----DGDLRRVSLRLRNVGPPVDPVTE
Mvan_2410|M.vanbaalenii_PYR-1 VDGGRLVRTRRSLRAALSMSVEP----DDGLLRLTVEVRNTAAPVA----
TH_1949|M.thermoresistible__bu VTGGRLVRSRRALHGELTVTAEPGTAPGRELRVVEVTVRNTGTPTTD---
MSMEG_2715|M.smegmatis_MC2_155 VEGGRLVRTRRALCGELLLRAEP----DGDLFRVTMTVRNRAAPVED---
: ****** *. : . * : : . * : : :** ....
MAV_2686|M.avium_104 -KDSTIARSLIGTHIIAEAIGGEFISLLETPEHAAAAAARCRQHRCFPVL
MMAR_1865|M.marinum_M GKDSVIARSMIGSHMIAEVIGGQFVSLLEPAPAAADAVSRCTQHRCFPVL
Mvan_2410|M.vanbaalenii_PYR-1 DKDEAIATSLIGTHLLLEVDGGEFVSLLEPPDPAAAAAGRCRQHRCFPVL
TH_1949|M.thermoresistible__bu -RDDAIARSLIGAHVIARTGDGEFVSLLDPPAHAAAAAASCRQHRCFPVL
MSMEG_2715|M.smegmatis_MC2_155 -RQSATAVSFLGTHLIAEIDDGEFVSLLEPDDRAAEAVSRCERHRCFPVL
::.. * *::*:*:: . .*:*:***:. ** *.. * :*******
MAV_2686|M.avium_104 AGALGEHDLLLVSPIILYDHPEVAEQSEGALYDSTEIDEILTLRVMTMTD
MMAR_1865|M.marinum_M AGPPGTQDLLLISPIILYDHPEVAEQSNTALYDCTEIDEILTLRVMTMTD
Mvan_2410|M.vanbaalenii_PYR-1 AGPPGAHHLMLVSPIILYDHPEIAEQSKGDLYDACEIDEILTLRIMTMTE
TH_1949|M.thermoresistible__bu AGPPGTRDLLLVSPIILYDHPEIAEQSRTSLYDSTEIDEILTLRVMTLTD
MSMEG_2715|M.smegmatis_MC2_155 AGPDGTRDLMLVSPIILYDHPQIAAQSRGALYDSCEIDEILTLRVMALTD
**. * :.*:*:*********::* **. ***. *********:*::*:
MAV_2686|M.avium_104 QEKAQARATDPLAAEIIDRCDAMSPEAMQRMHGVLRDPHTGYPASDTGLI
MMAR_1865|M.marinum_M EEKAQARATDPRAAQIIDQCDAMSPEAMARLHGVLRDPHA-----VSGLV
Mvan_2410|M.vanbaalenii_PYR-1 EEKAAARATDTRAAEIIDRCDSMSPEAMLGLHGVLRDPHA----AAPPLI
TH_1949|M.thermoresistible__bu EEKAVARATDPLAAQLIDRCDSMSPEDLLRLHGVLRDPHAG--APPPTLI
MSMEG_2715|M.smegmatis_MC2_155 EEKLLARATDPLAAQIIDRCDAMSAEAMLDLHGVLRTTDDE----RSSLL
:** *****. **::**:**:**.* : :***** .. . *:
MAV_2686|M.avium_104 PEIPDGLGWWDPEADNAVRPDVDAILINGVRVARGSRVRLHPGRRADAQD
MMAR_1865|M.marinum_M PELPEGVDWWDPMADNAVRPELDGVLVNGVRVARGSRVRLHPRRNADAQD
Mvan_2410|M.vanbaalenii_PYR-1 PEIPADVDWWDPEADNAVHPESDAVLVNGARVARGSRVRLHPSRRADAQD
TH_1949|M.thermoresistible__bu PEVPDGIDWWDPSADTAVDPGLDAVLVDGVAVRRGSRVRLRPSRRADAQD
MSMEG_2715|M.smegmatis_MC2_155 PEVPDGVPWWDPLADNAVDPAVDAVLVNGVAVSRGCRVRLHPSRRADAQD
**:* .: **** **.** * *.:*::*. * **.****:* *.*****
MAV_2686|M.avium_104 LFVDGKSARVTSVHEDVDGNQHVGVVVDDDPDADLHDWYGRYLYFAPDEV
MMAR_1865|M.marinum_M IFVAGKIARVTSVHEDVEGNRHVGVVVEDDPAADLHDWYGRYLYFSPDEV
Mvan_2410|M.vanbaalenii_PYR-1 LFYADRIARVMSVHADVEGGSHIGVVLDDDPAADLHDWYGRYLYFAPDEV
TH_1949|M.thermoresistible__bu LFFAGRTAQVTSVHQDVDGATHIGVVLDDDPAAELHDWYGRYLYFAPDEV
MSMEG_2715|M.smegmatis_MC2_155 LFFDNRIAWVASIHEDVDGERHVGVVLDDDPAADLHDWYGRYLYFAPDEI
:* .: * * *:* **:* *:***::*** *:***********:***:
MAV_2686|M.avium_104 EPLEAH--PTGTERNPTWR-
MMAR_1865|M.marinum_M EPLESE--QTSNERSSIWRS
Mvan_2410|M.vanbaalenii_PYR-1 EPL--------VERSQ----
TH_1949|M.thermoresistible__bu EPLTDTREDTREERSPTWK-
MSMEG_2715|M.smegmatis_MC2_155 EPI-----------------
**: