For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGSLSPWHWAILAVVVILLFGAKKLPDAARSLGKSMRIFKSELREMQSENKTETSALGAQSESSAANPTP VQSQRVDPPAPSEQGHSEARPAS
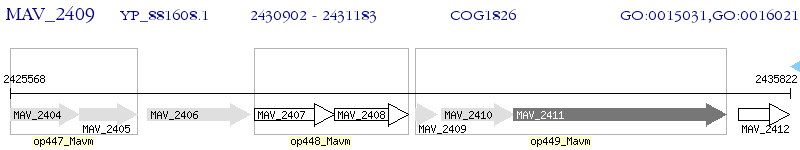
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2409 | tatA | - | 100% (93) | twin argininte translocase protein A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2121c | tatA | 6e-32 | 72.83% (92) | twin arginine translocase protein A |
| M. gilvum PYR-GCK | Mflv_3093 | tatA | 7e-21 | 51.06% (94) | twin arginine-targeting protein translocase |
| M. tuberculosis H37Rv | Rv2094c | tatA | 6e-32 | 72.83% (92) | twin arginine translocase protein A |
| M. leprae Br4923 | MLBr_01331 | tatA | 8e-34 | 73.91% (92) | twin arginine translocase protein A |
| M. abscessus ATCC 19977 | MAB_2187 | - | 5e-17 | 48.84% (86) | Sec-independent protein translocase protein TatA/E |
| M. marinum M | MMAR_3080 | tatA | 5e-35 | 77.17% (92) | sec-independent protein translocase membrane- bound |
| M. smegmatis MC2 155 | MSMEG_3887 | tatA | 9e-22 | 58.33% (84) | twin arginine-targeting protein translocase, TatA/E family |
| M. thermoresistible (build 8) | TH_0501 | - | 2e-26 | 59.78% (92) | PROBABLE SEC-INDEPENDENT PROTEIN TRANSLOCASE MEMBRANE-BOUND |
| M. ulcerans Agy99 | MUL_2326 | tatA | 4e-35 | 77.17% (92) | twin arginine translocase protein A |
| M. vanbaalenii PYR-1 | Mvan_3442 | tatA | 2e-20 | 53.26% (92) | twin arginine-targeting protein translocase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3093|M.gilvum_PYR-GCK -------MGGLQPWHWLIVIAVFVLLFGAKKLPDAARSLGKSMRIFKSEI
Mvan_3442|M.vanbaalenii_PYR-1 -------MGGLQPWHWVIVIAVFVLLFGAKKLPDAARSLGKSMRIFKSEI
MSMEG_3887|M.smegmatis_MC2_155 -------MGGLQPWHWVIVIAVFVLLFGAKKLPDAARSLGKSMRIFKSEI
MAB_2187|M.abscessus_ATCC_1997 ----MTTLGGLQPMHWVIVIVVFALLFGAKKLPDLARSLGKSLRIFKSEV
Mb2121c|M.bovis_AF2122/97 -------MGSLSPWHWAILAVVVIVLFGAKKLPDAARSLGKSLRIFKSEV
Rv2094c|M.tuberculosis_H37Rv -------MGSLSPWHWAILAVVVIVLFGAKKLPDAARSLGKSLRIFKSEV
MMAR_3080|M.marinum_M -------MGSLSPWHWAILAVVVIVLFGAKKLPDAARSLGKSMRIFKSEM
MUL_2326|M.ulcerans_Agy99 -------MGSLSPWHWAILAVVVIVLFGAKKLPDAARSLGKSMRIFKSEM
MAV_2409|M.avium_104 -------MGSLSPWHWAILAVVVILLFGAKKLPDAARSLGKSMRIFKSEL
MLBr_01331|M.leprae_Br4923 MSGGKFKVGSLSPWHWVVLVVVVVLLFGAKKLPDAARSLGKSMRIFKSEL
TH_0501|M.thermoresistible__bu -------VGALQPWHWLIVIIAFILLFGAKKLPDAARSLGRSMRIFKSEL
:*.*.* ** :: .. :********* *****:*:******:
Mflv_3093|M.gilvum_PYR-GCK KEMQSDSNAAKS------DQPEQITSERVVVDPSTQSTS--SNSDKRPA-
Mvan_3442|M.vanbaalenii_PYR-1 KEMQADSQSAKP------DQPAAPT--QIASERVPEAEP--SSPDKRPA-
MSMEG_3887|M.smegmatis_MC2_155 KEMQAES---KG------DEPKPAT--PIASERVDTTAPEQQSTDRHTA-
MAB_2187|M.abscessus_ATCC_1997 KELQNEGNAAKAS-----DTPQAVEQPQVPPTVAVEAPPIESTPGHKPTA
Mb2121c|M.bovis_AF2122/97 RELQNENKAEAS-----IET----PTPVQSQRVDPSAASGQDSTEARPA-
Rv2094c|M.tuberculosis_H37Rv RELQNENKAEAS-----IET----PTPVQSQRVDPSAASGQDSTEARPA-
MMAR_3080|M.marinum_M REMQSETKAEPSA----IETNTANPTPVQSQRIDPAAATGQDQTEARPA-
MUL_2326|M.ulcerans_Agy99 REMQSETKAEPSA----IETNTANPTPVQSQRIDPAAATGQDQTEARPA-
MAV_2409|M.avium_104 REMQSENKTETSALGAQSESSAANPTPVQSQRVDPPAPSEQGHSEARPAS
MLBr_01331|M.leprae_Br4923 REMQTENQAQASA----LETPMQNPTVVQSQRVVPPWSTEQDHTEARPA-
TH_0501|M.thermoresistible__bu KELQSDSQPSATT-----PPPAAPATPVDAERVDTPADSQQQHSDPRPA-
:*:* : . . :.: