For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDGDVRGSDTDKGAGAPNEPDEAALAAMSQQELVTLGGKLDGVETVFKEPRWPVEGTKAEKRAERGVALW LLLGGGFGLALLLIFLFWPWEYKPKEAAGSFLYDLATPLYGLTFGMSILSIGIGTILYQKRFIPEEISIQ ERHDGASREIDRKTVVANLADAYNGSTIGRRKMVGLSLGVGLGAFGLSTLVAFAGGLIKNPWKPVVPTAN GKKAVLWTSGWTPRYPGETIYLARATGTYTGAPFVKMRPEDLDAGGMETVFPWRESDGDGTTPESQEKLR AINQGVRNPVMLIRIRPTDMPRVVKRQGQESFNFGEFFAYTKVCSHLGCPASLYEEQSYRILCPCHQSQF DALHFAKPIFGPAARALAQLPITIDTNGYLVANGDFVEPVGPAFWERTTT*
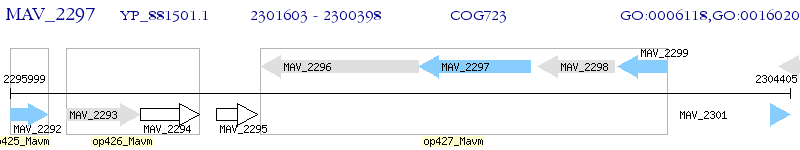
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2297 | - | - | 100% (401) | putative ubiquinol-cytochrome c reductase, iron-sulfur subunit QcrA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2218 | qcrA | 0.0 | 83.42% (392) | Rieske iron-sulfur protein QcrA |
| M. gilvum PYR-GCK | Mflv_2954 | - | 0.0 | 76.52% (396) | Rieske (2Fe-2S) domain-containing protein |
| M. tuberculosis H37Rv | Rv2195 | qcrA | 0.0 | 83.42% (392) | Rieske iron-sulfur protein QcrA |
| M. leprae Br4923 | MLBr_00880 | qcrA | 0.0 | 82.17% (387) | putative Rieske iron-sulphur reductase component |
| M. abscessus ATCC 19977 | MAB_1967c | - | 1e-168 | 72.38% (391) | ubiquinol-cytochrome c reductase iron-sulfur subunit (Rieske |
| M. marinum M | MMAR_3239 | qcrA | 0.0 | 83.72% (393) | Rieske iron-sulfur protein QcrA |
| M. smegmatis MC2 155 | MSMEG_4262 | - | 0.0 | 78.17% (394) | ubiquinol-cytochrome c reductase iron-sulfur subunit |
| M. thermoresistible (build 8) | TH_1043 | qcrA | 0.0 | 78.17% (394) | Probable Rieske iron-sulfur protein QcrA |
| M. ulcerans Agy99 | MUL_3552 | qcrA | 0.0 | 83.72% (393) | Rieske iron-sulfur protein QcrA |
| M. vanbaalenii PYR-1 | Mvan_3559 | - | 0.0 | 78.25% (400) | Rieske (2Fe-2S) domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2954|M.gilvum_PYR-GCK ----------------------MSQGPDDRNGPEKGTDVPGHKGEPGQPT
Mvan_3559|M.vanbaalenii_PYR-1 ----------------------MSDGHDD----VKGTDAPGQTGAPGQPT
MSMEG_4262|M.smegmatis_MC2_155 ----------------MDRIASMSQDSPD----IKGTDAPGQTGVPGQPT
TH_1043|M.thermoresistible__bu ----------------------MT---DD----VKGTDTPGQLGVPGQPT
Mb2218|M.bovis_AF2122/97 MSRADDDAVGVPPTCGGRSDEEERRIVPGPNPQDGAKDGAKATAVPREPD
Rv2195|M.tuberculosis_H37Rv MSRADDDAVGVPPTCGGRSDEEERRIVPGPNPQDGAKDGAKATAVPREPD
MMAR_3239|M.marinum_M -----------------------------------MSDGSTETGVSREPD
MUL_3552|M.ulcerans_Agy99 -----------------------------------MSDGSTETGVSREPD
MAV_2297|M.avium_104 ----------------------------MSDGDVRGSDTDKGAGAPNEPD
MLBr_00880|M.leprae_Br4923 ----------------------------MS------LNPKDGAGASRELD
MAB_1967c|M.abscessus_ATCC_199 -------------------------------------MSVSDADKPEIPS
.
Mflv_2954|M.gilvum_PYR-GCK DAELAKMSQEELLELGGRLDGVEIVHKEPRWPIPGTKAEKRAERTVAYWF
Mvan_3559|M.vanbaalenii_PYR-1 DAELAKMSQEELLELGGKLDGVEIVYKEPRWPVPGTKAEKRAERTVAYWF
MSMEG_4262|M.smegmatis_MC2_155 DAELAEMSREELVKLGGKIDGVETIFKEPRWPVPGTKAEKRTERLVAYWL
TH_1043|M.thermoresistible__bu EAELAQMSREELLELGGKLDGVQTVFKEPRWPVPGTKAEKRAERQVAFWL
Mb2218|M.bovis_AF2122/97 EAALAAMSNQELLALGGKLDGVRIAYKEPRWPVEGTKAEKRAERSVAVWL
Rv2195|M.tuberculosis_H37Rv EAALAAMSNQELLALGGKLDGVRIAYKEPRWPVEGTKAEKRAERSVAVWL
MMAR_3239|M.marinum_M EAALAEMSQQELLELGGSIDGVRIAHKEPRWPVEGTKAEKRAERGVALWL
MUL_3552|M.ulcerans_Agy99 EAALAEMSQQELLELGGSIDGVRIAHKEPRWPVEGTKAEKRAERGVALWL
MAV_2297|M.avium_104 EAALAAMSQQELVTLGGKLDGVETVFKEPRWPVEGTKAEKRAERGVALWL
MLBr_00880|M.leprae_Br4923 ETTQGTMSREELVALGGRLDGVNTVYKEQRWPIEGTKAEKRTERMVTFWL
MAB_1967c|M.abscessus_ATCC_199 DEQLAAMSRDELVKLGTNLDGVEIVYREPRWPVEGTKAEKRAERLVALWF
: . **.:**: ** :***. .:* ***: *******:** *: *:
Mflv_2954|M.gilvum_PYR-GCK LLAGFSGLALLLIFLFWPWEYKPYGSGGEFLYSLTTPLYGLTFGLSVLAI
Mvan_3559|M.vanbaalenii_PYR-1 LLAGFSGLALLLIFLFWPWEYKPFGDEGEFLYSLTTPLYGLTFGLSVLAI
MSMEG_4262|M.smegmatis_MC2_155 MLGGLSGLALLLVFLFWPWEYQPFGSEGEFLYSLATPLYGLTFGLSILSI
TH_1043|M.thermoresistible__bu MLGGFSGLALLLIFLFWPWEYRPLGDEGAWIYNLTTPLYGLTFGLSILSI
Mb2218|M.bovis_AF2122/97 LLGGVFGLALLLIFLFWPWEFKAADGESDFIYSLTTPLYGLTFGLSILSI
Rv2195|M.tuberculosis_H37Rv LLGGVFGLALLLIFLFWPWEFKAADGESDFIYSLTTPLYGLTFGLSILSI
MMAR_3239|M.marinum_M LLGGFFGLALLLVFLFWPWEYKPSGDKGNLLYSLATPLYGLTFGMSVLAI
MUL_3552|M.ulcerans_Agy99 LLGGFFGLALLLVFLFWPWEYKPSGDKGNLLYSLATPLYGLTFGMSVLAI
MAV_2297|M.avium_104 LLGGGFGLALLLIFLFWPWEYKPKEAAGSFLYDLATPLYGLTFGMSILSI
MLBr_00880|M.leprae_Br4923 LLGGVLGLTLLLIFLFWPWEYKPKEASGSFLYTIATPLYGLTFGLSILAI
MAB_1967c|M.abscessus_ATCC_199 ALGGVFGLALLGVFLFWPWEYKPFGDPGNALYNLATPLYGLTLGLSVLSL
*.* **:** :*******::. . :* ::*******:*:*:*::
Mflv_2954|M.gilvum_PYR-GCK GVGAVLFQKKFIPEEISIQDRHDG-RSPELHRKTIAANLGDALDGSTIKR
Mvan_3559|M.vanbaalenii_PYR-1 GVGAVLFQKKFIPEEISIQERHDG-RSPELHRKTVAANLGDALDGSTIKR
MSMEG_4262|M.smegmatis_MC2_155 GIGAVLFQKKFIPEEISVQDRHDG-RSPEVHRKTVAANLTDALEGSTLKR
TH_1043|M.thermoresistible__bu GIGAVLYQKKFIPHEISIQERHDG-RSPEIHRKTIVANLTDALEGSTIKR
Mb2218|M.bovis_AF2122/97 AIGAVLYQKRFIPEEISIQERHDG-ASREIDRKTVVANLTDAFEGSTIRR
Rv2195|M.tuberculosis_H37Rv AIGAVLYQKRFIPEEISIQERHDG-ASREIDRKTVVANLTDAFEGSTIRR
MMAR_3239|M.marinum_M AIGAVLFQKRFIPEEITIQDRHDG-ASREVDRKTVVANLADAFEASTIGR
MUL_3552|M.ulcerans_Agy99 AIGAVLFQKRFIPEEITIQDRHDG-ASREVDRKTVVANLADAFEASTIGR
MAV_2297|M.avium_104 GIGTILYQKRFIPEEISIQERHDG-ASREIDRKTVVANLADAYNGSTIGR
MLBr_00880|M.leprae_Br4923 AIGAVQFQKRFIPEEISIQERHDG-ASPEIDRKTVVANLTDALESSTIRR
MAB_1967c|M.abscessus_ATCC_199 GIGAVLYTKKFIPQEISIQDRHDGPGSAEVDRKTIVAQLQDTLDTSTLPR
.:*:: : *:***.**::*:**** * *:.***:.*:* *: : **: *
Mflv_2954|M.gilvum_PYR-GCK RKMIGLSLGIGFGAFGLGTAVAFIGGLIKNPWKPVVPTAEGMKAVLWTSG
Mvan_3559|M.vanbaalenii_PYR-1 RKMIGLSLGIGFGAFGLGTAVAFIGGLIKNPWKPVVPTAEGPKAVLWTSG
MSMEG_4262|M.smegmatis_MC2_155 RKVIGLSLGIGLGAFGAGTLVAFIGGLIKNPWKPVVPTAEGKKAVLWTSG
TH_1043|M.thermoresistible__bu RKVIGLSLGLGLGAFGLGTLVAFVGGLVKNPWKPVVPTAEGKKAELWTSG
Mb2218|M.bovis_AF2122/97 RKLIGLSFGVGMGAFGLGTLVAFAGGLIKNPWKPVVPTAEGKKAVLWTSG
Rv2195|M.tuberculosis_H37Rv RKLIGLSFGVGMGAFGLGTLVAFAGGLIKNPWKPVVPTAEGKKAVLWTSG
MMAR_3239|M.marinum_M RKMVGASLGVGLGAFGLGTLVAFAGGLIKNPWKPVVPTADGKKAVLWTSG
MUL_3552|M.ulcerans_Agy99 RKMVGASLGVGLGAFGLGTLVAFAGGLIKNPWKPVVPTADGKKAVLWTSG
MAV_2297|M.avium_104 RKMVGLSLGVGLGAFGLSTLVAFAGGLIKNPWKPVVPTANGKKAVLWTSG
MLBr_00880|M.leprae_Br4923 RKLIGLSLGVGLGAFGLSTLVTFIGGFIKNPWKPVVPTANGMKAMLWTSG
MAB_1967c|M.abscessus_ATCC_199 RKMIVRALGFGVGAMGAGTAVAFIGGMIKNPWARVVPTANGKQPVLLTSG
**:: ::*.*.**:* .* *:* **::**** *****:* :. * ***
Mflv_2954|M.gilvum_PYR-GCK WTPRFEGETIFLARATGRPGNSPFVKMRPEDLDAGGMETVYPWRESDGDG
Mvan_3559|M.vanbaalenii_PYR-1 WTPRFHGETIYLARATGKPGESPYVKMRPEDLDAGGMETVYPWRESDGDG
MSMEG_4262|M.smegmatis_MC2_155 WTPRFKGETIYLARATGRPGESPFVKMRPEDIDAGGMETVFPWRESDGDG
TH_1043|M.thermoresistible__bu WTPRYPGETIYLVRSTGAVGTAPFQKVRPEDIDAGGMETVFPWRESDGDG
Mb2218|M.bovis_AF2122/97 WTPRYQGETIYLARATGTEDGPPFIKMRPEDIDAGGMETVFPWRESDGDG
Rv2195|M.tuberculosis_H37Rv WTPRYQGETIYLARATGTEDGPPFIKMRPEDMDAGGMETVFPWRESDGDG
MMAR_3239|M.marinum_M WTPRFHGETIYLARATGLGDGPPYTKMRPEDLDAGGMETVFPWRESDGDG
MUL_3552|M.ulcerans_Agy99 WTPRFHGETIYLARATGLGDGPPYAKMRPEDLDAGGMETVFPWRESDGDG
MAV_2297|M.avium_104 WTPRYPGETIYLARATGTYTGAPFVKMRPEDLDAGGMETVFPWRESDGDG
MLBr_00880|M.leprae_Br4923 WTPRYHGETIYLARATGSHSGAPFIRMRPEDVDAGGMETVFPWRESDGDG
MAB_1967c|M.abscessus_ATCC_199 WTPRYHGETIYLGRAVGSSSN-VLLKIRPEDIDAGGMETVFPWRESDGDG
****: ****:* *:.* ::****:********:*********
Mflv_2954|M.gilvum_PYR-GCK TTVESEHHLYDIVMGVRNPVMLIRIKPGDMSRVVKRRGQESFNFGELFAY
Mvan_3559|M.vanbaalenii_PYR-1 TTVESEHHLFEIAMGVRNPVMLIRIKPADMKRVVKRRGQESFNFGELFAY
MSMEG_4262|M.smegmatis_MC2_155 TTVESEHKLTEIAMGVRNPVMLIRIKPADMHRVIKRKGQESFNFGELFAY
TH_1043|M.thermoresistible__bu TTPESAHKLTEIAMGVRNPVMLIRIRPQDMPRVVKRKGQESFNYGEFFAY
Mb2218|M.bovis_AF2122/97 TTVESHHKLQEIAMGIRNPVMLIRIKPSDLGRVVKRKGQESFNFGEFFAF
Rv2195|M.tuberculosis_H37Rv TTVESHHKLQEIAMGIRNPVMLIRIKPSDLGRVVKRKGQESFNFGEFFAF
MMAR_3239|M.marinum_M TTVESHEKLQRISMGVRNPVMLIRIKPTDMHRVVKRQGQESFNFGELFAF
MUL_3552|M.ulcerans_Agy99 TTVESHEKLQRISMGVRNPVMLIRIKPTDMHRVVKRQGQESFNFGELFAF
MAV_2297|M.avium_104 TTPESQEKLRAINQGVRNPVMLIRIRPTDMPRVVKRQGQESFNFGEFFAY
MLBr_00880|M.leprae_Br4923 TTMESHEKLITIKMGIRNPVMLIRIRPSDMPRVVKRRGQENFNFGEFFAF
MAB_1967c|M.abscessus_ATCC_199 TTVESHEKLSHIQMGIRNPVMLIRVRPTDMPKVVKRKGQENFNYGDLFAY
** ** .:* * *:********::* *: :*:**:***.**:*::**:
Mflv_2954|M.gilvum_PYR-GCK TKVCSHLGCPSSLYEQQTYRILCPCHQSQFDALHFAKPIFGPAARALAQL
Mvan_3559|M.vanbaalenii_PYR-1 TKVCSHLGCPSSLYEQQTYRILCPCHQSQFDALHFAKPIFGPAARALAQL
MSMEG_4262|M.smegmatis_MC2_155 TKVCSHLGCPSSLYEQQTYRILCPCHQSQFDALEFAKPIFGPAARALAQL
TH_1043|M.thermoresistible__bu TKVCSHLGCPASLYEQQTHLLLCPCHQSQFDLLHYARPVFGPAARALAQL
Mb2218|M.bovis_AF2122/97 TKVCSHLGCPSSLYEQQSYRILCPCHQSQFDALHFAKPIFGPAARALAQL
Rv2195|M.tuberculosis_H37Rv TKVCSHLGCPSSLYEQQSYRILCPCHQSQFDALHFAKPIFGPAARALAQL
MMAR_3239|M.marinum_M TKVCSHLGCPSSLYEQQSYRILCPCHQSQFDALHFAKPIFGPAARALAQL
MUL_3552|M.ulcerans_Agy99 TKVCSHLGCPSSLYEQQSYRILCPCHQSQFDALHFAKPIFGPAARALAQL
MAV_2297|M.avium_104 TKVCSHLGCPASLYEEQSYRILCPCHQSQFDALHFAKPIFGPAARALAQL
MLBr_00880|M.leprae_Br4923 TKVCSHLGCPSSLYEQQSYRILCPCHQSQFDALQYAKPIFGPAARALAQL
MAB_1967c|M.abscessus_ATCC_199 TKICSHLGCPTSLFEQQTYRILCPCHQSQFDALHFAKPIFGPAARALAQL
**:*******:**:*:*:: :********** *.:*:*:***********
Mflv_2954|M.gilvum_PYR-GCK PITIDQDGYLVANGDFIEPVGPAFWERKS-
Mvan_3559|M.vanbaalenii_PYR-1 PITIDQDGYLVANGDFIEPVGPAFWERKS-
MSMEG_4262|M.smegmatis_MC2_155 PITIDEDGYLVANGDFVEPVGPAFWERKS-
TH_1043|M.thermoresistible__bu PITVNEDGYLVANGDFIEPVGPAFWERKS-
Mb2218|M.bovis_AF2122/97 PITIDTDGYLVANGDFVEPVGPAFWERTTT
Rv2195|M.tuberculosis_H37Rv PITIDTDGYLVANGDFVEPVGPAFWERTTT
MMAR_3239|M.marinum_M PITIDTEGYLVANGDFIEPVGPAFWERTTS
MUL_3552|M.ulcerans_Agy99 PITIDTEGYLVANGDFIEPVGPAFWERTTS
MAV_2297|M.avium_104 PITIDTNGYLVANGDFVEPVGPAFWERTTT
MLBr_00880|M.leprae_Br4923 PITIDTDGYLVANGDFVEPVGPAFWERTT-
MAB_1967c|M.abscessus_ATCC_199 PLTVDKDGYLVANGDFVEPVGPAFWERKS-
*:*:: :*********:**********.: