For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVQNESNAKTHGVILTEAAATKAKSLLDQEGRDDLALRIAVQPGGCAGLRYNLFFDDRSLDGDLTAEFG GVTLTVDRMSAPYVEGASIDFVDTIEKQGFTIDNPNATGSCACGDSFN
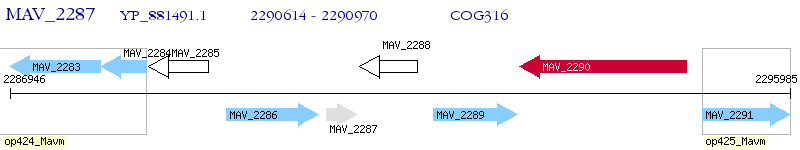
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2287 | - | - | 100% (118) | HesB/YadR/YfhF family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2227c | - | 7e-61 | 94.07% (118) | hypothetical protein Mb2227c |
| M. gilvum PYR-GCK | Mflv_2945 | - | 2e-54 | 80.80% (125) | iron-sulfur cluster assembly accessory protein |
| M. tuberculosis H37Rv | Rv2204c | - | 7e-61 | 94.07% (118) | hypothetical protein Rv2204c |
| M. leprae Br4923 | MLBr_00871 | - | 9e-62 | 94.07% (118) | hypothetical protein MLBr_00871 |
| M. abscessus ATCC 19977 | MAB_1957 | - | 2e-56 | 84.55% (123) | hypothetical protein MAB_1957 |
| M. marinum M | MMAR_3248 | - | 5e-63 | 96.61% (118) | hypothetical protein MMAR_3248 |
| M. smegmatis MC2 155 | MSMEG_4272 | - | 2e-58 | 88.98% (118) | HesB/YadR/YfhF family protein |
| M. thermoresistible (build 8) | TH_2400 | - | 8e-60 | 91.67% (120) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3561 | - | 2e-62 | 95.76% (118) | hypothetical protein MUL_3561 |
| M. vanbaalenii PYR-1 | Mvan_3568 | - | 2e-55 | 82.40% (125) | iron-sulfur cluster assembly accessory protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3248|M.marinum_M MTVQNESD-------AKTHGVILTEAAAAKAKSLLDQEGRDDLSLRIAVQ
MUL_3561|M.ulcerans_Agy99 MTVQNESD-------AKTHGVILTEAAAAKAKSLLDQEGRDDLSLRIAVQ
MAV_2287|M.avium_104 MTVQNESN-------AKTHGVILTEAAATKAKSLLDQEGRDDLALRIAVQ
Mb2227c|M.bovis_AF2122/97 MTVQNEPS-------AKTHGVILTEAAAAKAKSLLDQEGRDDLALRIAVQ
Rv2204c|M.tuberculosis_H37Rv MTVQNEPS-------AKTHGVILTEAAAAKAKSLLDQEGRDDLALRIAVQ
MLBr_00871|M.leprae_Br4923 MAVQNELS-------AKTHGVILTDVAATKAKSLLDQEGRDDLALRIAVQ
TH_2400|M.thermoresistible__bu MTVQDDSAT-----GTKTHGVILTEAAASKAKALLDQEGRDDLALRIAVQ
MSMEG_4272|M.smegmatis_MC2_155 MTVQDATA-------TETHGVTLTDAAAAKAKALLDQEGRDDLALRIAVQ
Mflv_2945|M.gilvum_PYR-GCK MTVQDESAVQAGQTTEAAHGVTLSDAAAAKAKALLDQEGRDDLALRIAVQ
Mvan_3568|M.vanbaalenii_PYR-1 MTVQDESAVQAGQTTEAQHGVTLSDAAAAKAKALLDQEGRDDLALRIAVQ
MAB_1957|M.abscessus_ATCC_1997 MTVQDESTATT--TDEGTHGVVLTDAAAAKAKALLDQEGRDDLSLRIAVQ
*:**: *** *::.**:***:**********:******
MMAR_3248|M.marinum_M PGGCAGLRYNLFFDDRALDGDLTAEFGGVTLTVDRMSAPYVEGASIDFVD
MUL_3561|M.ulcerans_Agy99 PGGCAGLRYNLFFDDRALDGDLTAEFGGVTLTVDRMSAPYVEGALIDFVD
MAV_2287|M.avium_104 PGGCAGLRYNLFFDDRSLDGDLTAEFGGVTLTVDRMSAPYVEGASIDFVD
Mb2227c|M.bovis_AF2122/97 PGGCAGLRYNLFFDDRTLDGDQTAEFGGVRLIVDRMSAPYVEGASIDFVD
Rv2204c|M.tuberculosis_H37Rv PGGCAGLRYNLFFDDRTLDGDQTAEFGGVRLIVDRMSAPYVEGASIDFVD
MLBr_00871|M.leprae_Br4923 PGGCAGLRYNLFFDDRTLDGDLTAEFGGVTLTVDRMSAPYVEGASIDFVD
TH_2400|M.thermoresistible__bu PGGCAGLRYNLFFDDRQLDGDLTAEFGGVTLTVDRMSAPYVEGATIDFVD
MSMEG_4272|M.smegmatis_MC2_155 PGGCAGLRYNLFFDDRTLDGDLTAEFGGVTLTVDRMSAPYVQGATIDFVD
Mflv_2945|M.gilvum_PYR-GCK PGGCAGLRYNLFFDDRTLDGDLTVDFSGVVLTVDRMSAPYVQGATIDFVD
Mvan_3568|M.vanbaalenii_PYR-1 PGGCAGLRYNLFFDDRSLDGDLTVDFGGVALTVDRMSAPYVQGATIDFVD
MAB_1957|M.abscessus_ATCC_1997 PGGCAGLRYQLFFDDRTLDGDLTVDFDGVTLTVDRMSAPYVQGASIDFVD
*********:****** **** *.:*.** * *********:** *****
MMAR_3248|M.marinum_M TIEKQGFTIDNPNATGSCACGDSFN
MUL_3561|M.ulcerans_Agy99 TIEKQGFTIDNPNATGSCACGDSFN
MAV_2287|M.avium_104 TIEKQGFTIDNPNATGSCACGDSFN
Mb2227c|M.bovis_AF2122/97 TIEKQGFTIDNPNATGSCACGDSFN
Rv2204c|M.tuberculosis_H37Rv TIEKQGFTIDNPNATGSCACGDSFN
MLBr_00871|M.leprae_Br4923 TIEKQGFTIDNPNANGSCACGDSFN
TH_2400|M.thermoresistible__bu TIEKQGFTIDNPNATGSCACGDSFN
MSMEG_4272|M.smegmatis_MC2_155 TIEKQGFTIDNPNATGSCACGDSFN
Mflv_2945|M.gilvum_PYR-GCK TIEKQGFTIDNPNATGSCACGDSFN
Mvan_3568|M.vanbaalenii_PYR-1 TIEKQGFTIDNPNATGSCACGDSFN
MAB_1957|M.abscessus_ATCC_1997 TIEKQGFTIDNPNATGSCACGDSFN
**************.**********