For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VALAVLFLIALAAGALGGLVGTGSSLVLLPALVMMYGPRIAVPVMGIAAVMANVARVAAWWRQIRWRPVL AYALPGTPAVVLGANTLLSIPQAVVDGLLGVFFLAMIPVRRVALARRWRIRLRHLAVAGVVVGFLTGLVL STGPLSVPVFTGYGLSGGAFLGSEAASALLLYIGKIGTFGQLGALSFAVVTRGLVIGIALMIGPFVARPL VRRTDQRHYALLIDGVLVVAAAGMFIATATH*
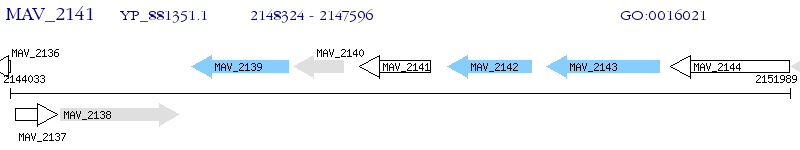
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2141 | - | - | 100% (242) | hypothetical protein MAV_2141 |
| M. avium 104 | MAV_3519 | - | 2e-06 | 28.09% (178) | hypothetical protein MAV_3519 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1361 | - | 6e-10 | 27.02% (248) | hypothetical protein Mflv_1361 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00468 | - | 7e-07 | 27.57% (185) | hypothetical protein MLBr_00468 |
| M. abscessus ATCC 19977 | MAB_4933 | - | 2e-06 | 23.26% (215) | hypothetical protein MAB_4933 |
| M. marinum M | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_4293 | - | 2e-07 | 28.27% (191) | PUTATIVE putative protein of unknown function DUF81 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5834 | - | 1e-08 | 29.84% (248) | hypothetical protein Mvan_5834 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1361|M.gilvum_PYR-GCK ----MIAVGVALGALIGVLLGLLGGGGSILAVPALVYGLGLGVEQAIPIS
Mvan_5834|M.vanbaalenii_PYR-1 ---MAVPLGLALGALIGVLLGLLGGGGSILAVPALVYGTGLSIGQAIPIS
TH_4293|M.thermoresistible__bu ----MIGLIIALAVLVGVALGLLGGGGSILMVPLLAYVAGMDAKAAIATS
MAV_2141|M.avium_104 --MVALAVLFLIALAAGALGGLVGTGSSLVLLPALVMMYGP----RIAVP
MAB_4933|M.abscessus_ATCC_1997 MDAPTLAVLLAAASAAGWVDAVVGGGGLILIPVLMLVFPGMAPATALGTN
MLBr_00468|M.leprae_Br4923 MLVSHLATIGLAGFGAGAINALVGSG--TLITFPTLVALGYSPVTSTMSN
: . * .::* * : *
Mflv_1361|M.gilvum_PYR-GCK LIVVAAASAVGVLPKIRARQVRWRMAGIFSAAGIPATLAGSAISRHLPES
Mvan_5834|M.vanbaalenii_PYR-1 LIVVAAASAVGALPRIRAGQVRWRMAGIFAAAGIPATVAGSAVSRHLPEP
TH_4293|M.thermoresistible__bu LLVVGVTSAVGAVSHARAGQVRWRTGLIFGAAGMAGAYAGGILARFIPGT
MAV_2141|M.avium_104 VMGIAAVMANVARVAAWWRQIRWRPVLAYALPGTPAVVLG-------ANT
MAB_4933|M.abscessus_ATCC_1997 KLTALSGTASAAIRLFPRTPLNWRVLVGAFFVAAAFASAGAYTASRLPVS
MLBr_00468|M.leprae_Br4923 AVGLVAGGVSATWGYRAELGDRWSRLRWQIPMSLTGAALGAFLLLHLPEK
: . . .* . . . * .
Mflv_1361|M.gilvum_PYR-GCK ALLIGFAAVMVVAGIRMLTDQSSTGTACEIREGEV-----NWRRCAPRSI
Mvan_5834|M.vanbaalenii_PYR-1 VLMIGFAVVMVVAGIRMLADQGHTGTACEIRGGQV-----DWRRCAPRSI
TH_4293|M.thermoresistible__bu VLLLGFAVMMAATAVAMLR--GRRNTATPDTGG-------DHRLPVPRIL
MAV_2141|M.avium_104 LLSIPQAVVDGLLGVFFLAMIPVRRVALARR----------WRIRLRHLA
MAB_4933|M.abscessus_ATCC_1997 VFKPVVLVLLVAVGVFVATRPQFGTTTEGTART--------KTATLTALT
MLBr_00468|M.leprae_Br4923 VFTAIVPMLLVLALVLVVAGPRIQSWTRCRAKAAVNRAEHTTPAQMAMLA
: : : . :
Mflv_1361|M.gilvum_PYR-GCK GAGLVVGLLTGLFGVGG--GFLIIPALVVVLGVEMSTAIGTSLLIIIANS
Mvan_5834|M.vanbaalenii_PYR-1 GAGLLVGVLTGLFGVGG--GFLIIPALVVVLGIEMSTAIGTSLLIIVANS
TH_4293|M.thermoresistible__bu ADGLVVGLVTGLVGAGG--GFLVVPALALLGGLSMPAAVGTSLVVIAMKS
MAV_2141|M.avium_104 VAGVVVGFLTGLVLSTGPLSVPVFTGYGLSGGAFLGSEAASALLLYIGK-
MAB_4933|M.abscessus_ATCC_1997 IAAVAIAFYDGIMGPGT--GTFLIITLTVAAGMTFLESSATAKVLNSGTN
MLBr_00468|M.leprae_Br4923 LGTFAVSVYGGYFTAGQGILLVSLMSILLPESMQRINAAKNLLSMLVNLT
. :.. * . . : . . :
Mflv_1361|M.gilvum_PYR-GCK VAGIFSHLQSISVDWQVTGLFV-AAAMVTSLIAGYVGTKTDTERLQRWFA
Mvan_5834|M.vanbaalenii_PYR-1 LAGLVSHLDAVGGNWSITAAFV-GAAMATSLVAGHFGTKVDTDRLQRWFA
TH_4293|M.thermoresistible__bu FAGLAGYLTSVQIDWPVALAVT-VAAVAGALLGSRLTALVNPDTLRQAFG
MAV_2141|M.avium_104 -IGTFGQLGALSFAVVTRGLVIGIALMIGPFVARPLVRRTDQRHYALLID
MAB_4933|M.abscessus_ATCC_1997 VGALVVFASQGHVLWLLGLALG-LANIVGAQLGAHMALGRGAGFVRVVLL
MLBr_00468|M.leprae_Br4923 AAVGYTLVAFDRINWSAAGMIA-ASSLVGGLVGTRYARRLSPNVLRATIV
. : : :. . :
Mflv_1361|M.gilvum_PYR-GCK YLVFVVAGYVLIDTIFLR--------------------------------
Mvan_5834|M.vanbaalenii_PYR-1 YLVFAVAAYVLVDTIIFR--------------------------------
TH_4293|M.thermoresistible__bu WFVLTMSAVILAQEAHPAVGAAAAVLTAVAAALTFTCSRYGRCPLRRVAE
MAV_2141|M.avium_104 GVLVVAAAGMFIATATH---------------------------------
MAB_4933|M.abscessus_ATCC_1997 AVVVVMVGKLGYDMLR----------------------------------
MLBr_00468|M.leprae_Br4923 AIGLIGLVRLLAV-------------------------------------
. . :
Mflv_1361|M.gilvum_PYR-GCK ---------
Mvan_5834|M.vanbaalenii_PYR-1 ---------
TH_4293|M.thermoresistible__bu ALSSRPATA
MAV_2141|M.avium_104 ---------
MAB_4933|M.abscessus_ATCC_1997 ---------
MLBr_00468|M.leprae_Br4923 ---------