For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KPFSESERQEFLAGTHVAVLSVEATDGRPPAAVPIWYDYTPGGDIRIMTGASSRKARLIERAGKVTVVVQ REEPPYQYVVVEGTVVDATKPAPSDVQLAVAIRYLGEEGGRAFVQSLEGVEEVLFTIRPDRWLSADFTGD L*
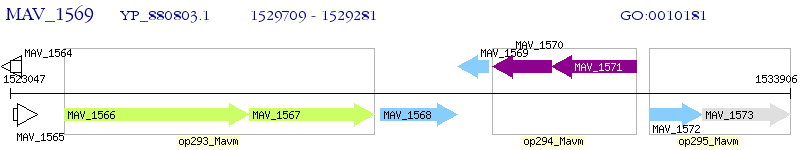
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1569 | - | - | 100% (142) | pyridoxamine 5'-phosphate oxidase family protein |
| M. avium 104 | MAV_1764 | - | 1e-12 | 34.09% (132) | pyridoxamine 5'-phosphate oxidase family protein |
| M. avium 104 | MAV_5242 | - | 6e-07 | 29.25% (106) | pyridoxamine 5'-phosphate oxidase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1695 | - | 6e-24 | 43.07% (137) | pyridoxamine 5'-phosphate oxidase-related, FMN-binding |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4748c | - | 4e-53 | 66.67% (141) | hypothetical protein MAB_4748c |
| M. marinum M | MMAR_0235 | - | 2e-07 | 31.07% (103) | pyridoxamine 5'-phosphate oxidase |
| M. smegmatis MC2 155 | MSMEG_5717 | - | 5e-26 | 45.52% (134) | pyridoxamine 5'-phosphate oxidase family protein |
| M. thermoresistible (build 8) | TH_2034 | - | 3e-24 | 43.70% (135) | pyridoxamine 5'-phosphate oxidase family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1188 | - | 1e-28 | 44.60% (139) | pyridoxamine 5'-phosphate oxidase-related, FMN-binding |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1569|M.avium_104 --------------MKPFSESERQEFLAGTHVAVLSVEATDGRPPAAVPI
MAB_4748c|M.abscessus_ATCC_199 -------------MPRKFTEAERHDFLAAKHVAVLSVATDDGRPPASVPI
Mflv_1695|M.gilvum_PYR-GCK -----MSPQLSSVRVMPLSKPEREEFLAEPHIAALSVANGADRGPLTVPI
MSMEG_5717|M.smegmatis_MC2_155 ---------------MALPKEEREQFLAEPHIAALSVYAGDKRGPLTVPI
TH_2034|M.thermoresistible__bu ---------------MALSKAEREEFLAEPRVAALSVYAGDRRGPLTLPI
Mvan_1188|M.vanbaalenii_PYR-1 --------------MATMTNDEREKYLAGLHVGVIAVER-PGRAPLSVPI
MMAR_0235|M.marinum_M MLLPLSAQPYTAPTDRDMLPSERREFVRTHRTCVFGYRRRNDGPGMSI-V
: **..:: : .:. :: :
MAV_1569|M.avium_104 WYDYTPGGDIRIMTGASSRKARLIERAGKVTVVVQREEPPYQYVVVEG-T
MAB_4748c|M.abscessus_ATCC_199 WYDYTPGGNIRINTGAGNRKARLIERAGAITLTVQHEEPPYQYVVVEG-T
Mflv_1695|M.gilvum_PYR-GCK WYQYSPGGELWFTTGAGSRKHRLIEAAGFVSLMVERVEPSVRYVAVDG-P
MSMEG_5717|M.smegmatis_MC2_155 WYQYSPGGEPWVLTGNGSRKHRLIESAGRFTLMVERLEPTVRYVAVDG-P
TH_2034|M.thermoresistible__bu WYQFQPGGQPWVLTGSGSRKHRLIEAAGHFSLMVERTEPTTRYVSVDG-P
Mvan_1188|M.vanbaalenii_PYR-1 WYGYEPGGEVLLWTEADSVKHRLIRDAGRFAITAQDEQPPYRYVTAEG-D
MMAR_0235|M.marinum_M YYIPTDDDELLVATMAGRGKARVVERDGKVSLCVLDERWPFTYLQVYADA
:* ..: . * . * *::. * .:: . . . *: . .
MAV_1569|M.avium_104 VVDATKPAPSDVQLAVAIRYLG--EEGGR--AFVQSLEGVEEVLFTIRPD
MAB_4748c|M.abscessus_ATCC_199 VVDTERPASAGPREEIAIRYLG--EEAGR--EFARSYDNTDTVLFTIKPD
Mflv_1695|M.gilvum_PYR-GCK VLRIE-PGTDDQLVEMTRRYLPP-EKVDGYLEFARREHG-ESVAVFVEAA
MSMEG_5717|M.smegmatis_MC2_155 VSRIE-PATDDHLVEVTKRYLAP-EKVDGYLEFARREHG-ESVVVFMTPE
TH_2034|M.thermoresistible__bu VSRIE-PGTDEMMAEMARRYLSA-ESADRYLQFARENLG-EHVAIYLEPR
Mvan_1188|M.vanbaalenii_PYR-1 VTAIG-PARDAEVRAIAVRYLG--EQEGG--QFTDENLTPTSVVIRMRPQ
MMAR_0235|M.marinum_M MLDPDRELAVDVMMAVAGRMSGQPLDAQARPSVEAMCERENRVVIRCRPY
: :: * . . * . .
MAV_1569|M.avium_104 --------------------RWLS-------ADFTGDL
MAB_4748c|M.abscessus_ATCC_199 --------------------RWFT-------ADFSGEL
Mflv_1695|M.gilvum_PYR-GCK --------------------HWLS-------SDLGSL-
MSMEG_5717|M.smegmatis_MC2_155 --------------------HWLS-------ADLGSV-
TH_2034|M.thermoresistible__bu --------------------HWLS-------ADLGSF-
Mvan_1188|M.vanbaalenii_PYR-1 --------------------RWLS-------TDYSK--
MMAR_0235|M.marinum_M STFATPPRHLHRNDQVEHLTHWVSGVVPWDAADPASSR
:*.: :*