For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPATETADKNTKSANSDTSGRVVRVTGPVVDVEFPRGSVPALFNALHAEITFEELAKTLTLEVAQHLGDN LVRTISLQPTDGLVRGVEVIDTGRSISVPVGQEVKGHVFNALGHCLDKPGYGEDFEHWSIHRKPPPFEEL EPRTEMLETGLKVVDLLTPYVRGGKIALFGGAGVGKTVLIQEMINRIARNFGGTSVFAGVGERTREGNDL WVELQEANVLKDTALVFGQMDEPPGTRMRVALSALTMAEWFRDEAGQDVLLFIDNIFRFTQAGSEVSTLL GRMPSAVGYQPTLADEMGELQERITSTRGRSITSMQAVYVPADDYTDPAPATTFAHLDATTELSRSVFSK GIFPAVDPLASSSTILDPGVVGEEHYRVAQEVIRILQRYKDLQDIIAILGIDELSEEDKQLVNRARRIER FLSQNMMAAEQFTGQPGSTVPLKETIEAFDRLTKGEFDHVPEQAFFLIGGLDDLAKKAESLGAKL
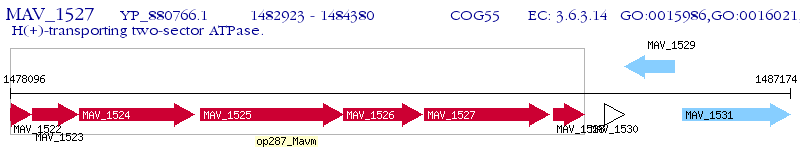
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1527 | atpD | - | 100% (485) | F0F1 ATP synthase subunit beta |
| M. avium 104 | MAV_1525 | atpA | 1e-22 | 25.11% (466) | F0F1 ATP synthase subunit alpha |
| M. avium 104 | MAV_1514 | rho | 8e-10 | 25.30% (249) | transcription termination factor Rho |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1342 | atpD | 0.0 | 94.80% (481) | F0F1 ATP synthase subunit beta |
| M. gilvum PYR-GCK | Mflv_2318 | - | 0.0 | 92.84% (475) | F0F1 ATP synthase subunit beta |
| M. tuberculosis H37Rv | Rv1310 | atpD | 0.0 | 94.80% (481) | F0F1 ATP synthase subunit beta |
| M. leprae Br4923 | MLBr_01145 | atpD | 0.0 | 92.16% (485) | F0F1 ATP synthase subunit beta |
| M. abscessus ATCC 19977 | MAB_1453 | - | 0.0 | 91.16% (475) | F0F1 ATP synthase subunit beta |
| M. marinum M | MMAR_4087 | atpD | 0.0 | 94.81% (482) | ATP synthase beta chain AtpD |
| M. smegmatis MC2 155 | MSMEG_4936 | atpD | 0.0 | 92.12% (482) | F0F1 ATP synthase subunit beta |
| M. thermoresistible (build 8) | TH_1558 | atpD | 0.0 | 91.49% (482) | PROBABLE ATP SYNTHASE BETA CHAIN ATPD |
| M. ulcerans Agy99 | MUL_3954 | atpD | 0.0 | 94.81% (482) | F0F1 ATP synthase subunit beta |
| M. vanbaalenii PYR-1 | Mvan_4328 | - | 0.0 | 91.29% (482) | F0F1 ATP synthase subunit beta |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4936|M.smegmatis_MC2_155 ----------MTATAEKT-AGRVVRITGPVVDVEFPRGSVPELFNALHAE
Mvan_4328|M.vanbaalenii_PYR-1 ----------MTATAEKT-AGRVVRITGPVVDVEFPRGSVPELFNALHAE
Mflv_2318|M.gilvum_PYR-GCK ----------MTSTEEKTTSGRVVRITGPVVDVEFPRGSVPELFNALHAD
Mb1342|M.bovis_AF2122/97 MTTTAEKTDRPGKPGSSDTSGRVVRVTGPVVDVEFPRGSIPELFNALHAE
Rv1310|M.tuberculosis_H37Rv MTTTAEKTDRPGKPGSSDTSGRVVRVTGPVVDVEFPRGSIPELFNALHAE
MMAR_4087|M.marinum_M ---MTVTAEKTDKSASSETSGRVVRVTGPVVDVEFPRGSVPELFNALHAK
MUL_3954|M.ulcerans_Agy99 ---MTVTAEKTDKSASSETSGRVVRVTGPVVDVEFPRGSVPELFNALHAK
MLBr_01145|M.leprae_Br4923 -MSTTKTTKMTVKTGSKGTSGRVVRVTGPVVDVEFPHGFVPELFNALNAK
MAV_1527|M.avium_104 -MPATETADKNTKSANSDTSGRVVRVTGPVVDVEFPRGSVPALFNALHAE
MAB_1453|M.abscessus_ATCC_1997 ---------MTAPTAN-KTAGRVVRVTGPVVDVEFPRDAVPPLFSALNAE
TH_1558|M.thermoresistible__bu ------MTATAEKETSKETTGRVVRVTGPVVDVEFPRGSVPDLFNALHAD
. :*****:**********:. :* **.**:*.
MSMEG_4936|M.smegmatis_MC2_155 ITFGALAKTLTLEVAQHLGDSLVRCISMQPTDGLVRGVEVTDTGASISVP
Mvan_4328|M.vanbaalenii_PYR-1 ISYKDLSKTLTLEVAQHLGDNLVRTISMQPTDGLVRGVEVTDTGTSISVP
Mflv_2318|M.gilvum_PYR-GCK ITYKELSKTLTLEVAQHLGDNLVRTISMQPTDGLVRGVEVTDTGNSISVP
Mb1342|M.bovis_AF2122/97 ITFESLAKTLTLEVAQHLGDNLVRTISLQPTDGLVRGVEVIDTGRSISVP
Rv1310|M.tuberculosis_H37Rv ITFESLAKTLTLEVAQHLGDNLVRTISLQPTDGLVRGVEVIDTGRSISVP
MMAR_4087|M.marinum_M IDFGSLAKTLTLEVAQHLGDSLVRTISLQPTDGLVRGTEVTDTGRPISVP
MUL_3954|M.ulcerans_Agy99 IDFGSLAKTLTLEVAQHLGDSLVRTISLQPTDGLVRGTEVTDTGRPISVP
MLBr_01145|M.leprae_Br4923 TTFSSLAKTLTLEVAQHLGDNLVRTISLQPTDGLVRGVEVTDTGNSISVP
MAV_1527|M.avium_104 ITFEELAKTLTLEVAQHLGDNLVRTISLQPTDGLVRGVEVIDTGRSISVP
MAB_1453|M.abscessus_ATCC_1997 ITYEAMAKTLTLEVAQHLGDNLVRTISMQPTDGLVRGVDVVSTGNTIAVP
TH_1558|M.thermoresistible__bu ITFGDLAKTLTLEVAQHLGDNMVRCISMQPTDGLVRGVDVRDTGRSISVP
: ::*************.:** **:*********.:* .** .*:**
MSMEG_4936|M.smegmatis_MC2_155 VGDGVKGHVFNALGDCLDDPGYGKDFEHWSIHRKPPAFSDLEPRTEMLET
Mvan_4328|M.vanbaalenii_PYR-1 VGDGVKGHVFNALGDCLDEPGYGKDFEHWSIHRKPPAFADLEPRTEMLET
Mflv_2318|M.gilvum_PYR-GCK VGDGVKGHVFNALGDCLDEPGYGKDFEHWSIHRKPPPFSELEPRTEMLET
Mb1342|M.bovis_AF2122/97 VGEGVKGHVFNALGDCLDEPGYGEKFEHWSIHRKPPAFEELEPRTEMLET
Rv1310|M.tuberculosis_H37Rv VGEGVKGHVFNALGDCLDEPGYGEKFEHWSIHRKPPAFEELEPRTEMLET
MMAR_4087|M.marinum_M VGESVKGHVFNALGDCLDEPGYGEDFEHWSIHRKPPAFEDLEPRTEMLET
MUL_3954|M.ulcerans_Agy99 VGESVKGHVFNALGDCLDEPGYGEDFEHWSIHRKPPAFEDLEPRTEMLET
MLBr_01145|M.leprae_Br4923 VGEGVKGHVFNALGYCLDEPGYGDEFEHWSIHRKPPSFEELEPRTEMLET
MAV_1527|M.avium_104 VGQEVKGHVFNALGHCLDKPGYGEDFEHWSIHRKPPPFEELEPRTEMLET
MAB_1453|M.abscessus_ATCC_1997 VGDGVKGHVFNALGNCLDEPGYGSDFEKWSIHRKPPAFDQLEPRTEMLET
TH_1558|M.thermoresistible__bu VGDGVKGHVFNALGDCLDQPGYGKDFEHWSIHRKPPAFSDLEPKTEMLET
**: ********** ***.****..**:********.* :***:******
MSMEG_4936|M.smegmatis_MC2_155 GLKVVDLLTPYVRGGKIALFGGAGVGKTVLIQEMINRIARNFGGTSVFAG
Mvan_4328|M.vanbaalenii_PYR-1 GLKVVDLLTPYVRGGKIALFGGAGVGKTVLIQEMINRIARNFGGTSVFAG
Mflv_2318|M.gilvum_PYR-GCK GLKVVDLLTPYVRGGKIALFGGAGVGKTVLIQEMINRIARNFGGTSVFAG
Mb1342|M.bovis_AF2122/97 GLKVVDLLTPYVRGGKIALFGGAGVGKTVLIQEMINRIARNFGGTSVFAG
Rv1310|M.tuberculosis_H37Rv GLKVVDLLTPYVRGGKIALFGGAGVGKTVLIQEMINRIARNFGGTSVFAG
MMAR_4087|M.marinum_M GLKVVDLLTPYVRGGKIALFGGAGVGKTVLIQEMINRIARNFGGTSVFAG
MUL_3954|M.ulcerans_Agy99 GLKVVDLLTPYVRGGKIALFGGAGVGKTVLIQEMINRIARNFGGTSVFAG
MLBr_01145|M.leprae_Br4923 GLKVVDLLTPYVRGGKIALFGGAGVGKTVLIQEMINRIARNFGGTSVFAG
MAV_1527|M.avium_104 GLKVVDLLTPYVRGGKIALFGGAGVGKTVLIQEMINRIARNFGGTSVFAG
MAB_1453|M.abscessus_ATCC_1997 GLKVVDLLTPYVRGGKIALFGGAGVGKTVLIQEMINRIARNFGGTSVFAG
TH_1558|M.thermoresistible__bu GLKVVDLLTPYVRGGKIALFGGAGVGKTVLIQEMINRIARNFGGTSVFAG
**************************************************
MSMEG_4936|M.smegmatis_MC2_155 VGERTREGNDLWVELADANVLKDTALVFGQMDEPPGTRMRVALSALTMAE
Mvan_4328|M.vanbaalenii_PYR-1 VGERTREGNDLWVELADANVLKDTALVFGQMDEPPGTRMRVALSALTMAE
Mflv_2318|M.gilvum_PYR-GCK VGERTREGNDLWVELEDANVLKDTALVFGQMDEPPGTRMRVALSALTMAE
Mb1342|M.bovis_AF2122/97 VGERTREGNDLWVELAEANVLKDTALVFGQMDEPPGTRMRVALSALTMAE
Rv1310|M.tuberculosis_H37Rv VGERTREGNDLWVELAEANVLKDTALVFGQMDEPPGTRMRVALSALTMAE
MMAR_4087|M.marinum_M VGERTREGNDLWVELQEANVLKDTALVFGQMDEPPGTRMRVALSALTMAE
MUL_3954|M.ulcerans_Agy99 VGERTREGNDLWVELQEANVLKDTALVFGQMDEPPGTRMRVALSALTMAE
MLBr_01145|M.leprae_Br4923 VGERTREGNDLWVELQEVNVLKDTALVFGQMDEPPGTRMRVALSALTMAE
MAV_1527|M.avium_104 VGERTREGNDLWVELQEANVLKDTALVFGQMDEPPGTRMRVALSALTMAE
MAB_1453|M.abscessus_ATCC_1997 VGERTREGNDLWVELADANVLKDTALVFGQMDEPPGTRMRVALSALTMAE
TH_1558|M.thermoresistible__bu VGERTREGNDLWVELGEADVLKDTALVFGQMDEPPGTRMRVALSALTMAE
*************** :.:*******************************
MSMEG_4936|M.smegmatis_MC2_155 FFRDEQGQDVLLFIDNIFRFTQAGSEVSTLLGRMPSAVGYQPTLADEMGE
Mvan_4328|M.vanbaalenii_PYR-1 FFRDEQGQDVLLFIDNIFRFTQAGSEVSTLLGRMPSAVGYQPTLADEMGE
Mflv_2318|M.gilvum_PYR-GCK FFRDEQQQDVLLFIDNIFRFTQAGSEVSTLLGRMPSAVGYQPTLADEMGE
Mb1342|M.bovis_AF2122/97 WFRDEQGQDVLLFIDNIFRFTQAGSEVSTLLGRMPSAVGYQPTLADEMGE
Rv1310|M.tuberculosis_H37Rv WFRDEQGQDVLLFIDNIFRFTQAGSEVSTLLGRMPSAVGYQPTLADEMGE
MMAR_4087|M.marinum_M WFRDEAGQDVLLFIDNIFRFTQAGSEVSTLLGRMPSAVGYQPTLADEMGE
MUL_3954|M.ulcerans_Agy99 WFRDEAGQDVLLFIDNIFRFTQAGSEVSTLLGRMPSAVGYQPTLADEMGE
MLBr_01145|M.leprae_Br4923 WFRDEASQDVLLFIDNIFRFTQAGSEVSTLLGRMPSAVGYQPTLADEMGE
MAV_1527|M.avium_104 WFRDEAGQDVLLFIDNIFRFTQAGSEVSTLLGRMPSAVGYQPTLADEMGE
MAB_1453|M.abscessus_ATCC_1997 YFRDEQGQDVLLFIDNIFRFTQAGSEVSTLLGRMPSAVGYQPTLADEMGE
TH_1558|M.thermoresistible__bu YFRDEQGQDVLLFIDNIFRFTQAGSEVSTLLGRMPSAVGYQPTLADEMGE
:**** *******************************************
MSMEG_4936|M.smegmatis_MC2_155 LQERITSTRGRSITSMQAVYVPADDYTDPAPATTFAHLDATTELSRAVFS
Mvan_4328|M.vanbaalenii_PYR-1 LQERITSTRGKSITSMQAVYVPADDYTDPAPATTFAHLDATTELSRAVFS
Mflv_2318|M.gilvum_PYR-GCK LQERITSTRGRSITSMQAVYVPADDYTDPAPATTFAHLDATTELSRTVFS
Mb1342|M.bovis_AF2122/97 LQERITSTRGRSITSMQAVYVPADDYTDPAPATTFAHLDATTELSRAVFS
Rv1310|M.tuberculosis_H37Rv LQERITSTRGRSITSMQAVYVPADDYTDPAPATTFAHLDATTELSRAVFS
MMAR_4087|M.marinum_M LQERITSTRGRSITSMQAVYVPADDYTDPAPATTFAHLDATTELSRAVFS
MUL_3954|M.ulcerans_Agy99 LQERITSTRGRSITSMQAVYVPADDYTDPAPATTFAHLDATTELSRAVFS
MLBr_01145|M.leprae_Br4923 LQERITSTRGRSITSMQAVYVPADDYTDPAPATTFAHLDATTELSRSVFA
MAV_1527|M.avium_104 LQERITSTRGRSITSMQAVYVPADDYTDPAPATTFAHLDATTELSRSVFS
MAB_1453|M.abscessus_ATCC_1997 LQERITSTRGRSITSMQAVYVPADDYTDPAPATTFAHLDATTELSRAVFS
TH_1558|M.thermoresistible__bu LQERITSTRGRSITSMQAVYVPADDYTDPAPATTFAHLDATTELSRAVFS
**********:***********************************:**:
MSMEG_4936|M.smegmatis_MC2_155 KGIFPAVDPLASSSTILDPAIVGDEHYRVAQEVIRILQRYKDLQDIIAIL
Mvan_4328|M.vanbaalenii_PYR-1 KGIFPAVDPLASSSTILDPAVVGDEHYRVAQEVIRILQRYKDLQDIIAIL
Mflv_2318|M.gilvum_PYR-GCK KGIFPAVDPLASSSTILDPAVVGDEHYRVAQEVIRILQRYKDLQDIIAIL
Mb1342|M.bovis_AF2122/97 KGIFPAVDPLASSSTILDPSVVGDEHYRVAQEVIRILQRYKDLQDIIAIL
Rv1310|M.tuberculosis_H37Rv KGIFPAVDPLASSSTILDPSVVGDEHYRVAQEVIRILQRYKDLQDIIAIL
MMAR_4087|M.marinum_M KGIFPAVDPLASSSTILDPGVVGDEHYRVAQEVIRILQRYKDLQDIIAIL
MUL_3954|M.ulcerans_Agy99 KGIFPAVDPLASSSTILDPGVVGDEHYRVAQEVIRILQRYKDLQDIIAIL
MLBr_01145|M.leprae_Br4923 KGIFPAVDPLASSSTILDPGIVGEEHYRVAQEVIRILQRYKDLQDIIAIL
MAV_1527|M.avium_104 KGIFPAVDPLASSSTILDPGVVGEEHYRVAQEVIRILQRYKDLQDIIAIL
MAB_1453|M.abscessus_ATCC_1997 KGIFPAVDPLASSSTILLPSVVGEEHYRVAQEVIRILQRYQDLQDIIAIL
TH_1558|M.thermoresistible__bu KGIFPAVDPLASSSTILDPSVVGDEHYRVAQEVIRILQRYKDLQDIIAIL
***************** *.:**:****************:*********
MSMEG_4936|M.smegmatis_MC2_155 GIDELSEEDKQLVNRARRIERFLSQNMMAAEQFTGQPGSTVPLKETIEAF
Mvan_4328|M.vanbaalenii_PYR-1 GIDELAEEDKQLVQRARRIERFLSQNMMAAEQFTGQPGSTVPLKETIEAF
Mflv_2318|M.gilvum_PYR-GCK GIDELAEEDKQLVQRARRLERFLSQNMMAAEQFTGQPGSTVPLKETIEAF
Mb1342|M.bovis_AF2122/97 GIDELSEEDKQLVNRARRIERFLSQNMMAAEQFTGQPGSTVPVKETIEAF
Rv1310|M.tuberculosis_H37Rv GIDELSEEDKQLVNRARRIERFLSQNMMAAEQFTGQPGSTVPVKETIEAF
MMAR_4087|M.marinum_M GIDELSEEDKQLVNRARRIERFLSQNMMAAEQFTGQPGSTVPVKETIEAF
MUL_3954|M.ulcerans_Agy99 GIDELSEEDKQLVNRARRIERFLSQNMMAAEQFTGQPGSTVPVKETIEAF
MLBr_01145|M.leprae_Br4923 GIDELSEEDKQLVNRARRIERFLSQNMMAAEQFTGQPGSTVPVKETIDAF
MAV_1527|M.avium_104 GIDELSEEDKQLVNRARRIERFLSQNMMAAEQFTGQPGSTVPLKETIEAF
MAB_1453|M.abscessus_ATCC_1997 GIDELSEEDKQLVGRARRIERFLSQNMMAAEQFTGQPGSTVPLKETIEAF
TH_1558|M.thermoresistible__bu GIDELSEEDKQLVNRARRIERFLSQNMMAAEQFTGQPGSTVPLKETIEAF
*****:******* ****:***********************:****:**
MSMEG_4936|M.smegmatis_MC2_155 DKLTKGEFDHLPEQAFFLIGGLDDLAKKAESLGAKL--------------
Mvan_4328|M.vanbaalenii_PYR-1 DKLTKGDFDHLPEQAFFLIGGLDDLAKKAESLGAKL--------------
Mflv_2318|M.gilvum_PYR-GCK DKLTKGEFDHLPEQAFFLIGGLDDLAKKAESLGAKL--------------
Mb1342|M.bovis_AF2122/97 DRLCKGDFDHVPEQAFFLIGGLDDLAKKAESLGAKL--------------
Rv1310|M.tuberculosis_H37Rv DRLCKGDFDHVPEQAFFLIGGLDDLAKKAESLGAKL--------------
MMAR_4087|M.marinum_M DRLCKGEFDHVPEQAFFLIGGLDDLAKKAESLGAKL--------------
MUL_3954|M.ulcerans_Agy99 DRLCKGEFDHVPEQAFFLIGGLDDLAKKAESLGAKL--------------
MLBr_01145|M.leprae_Br4923 DRLCKGEFDHVPEQAFFLIGGLDDLTKKAESLGAKL--------------
MAV_1527|M.avium_104 DRLTKGEFDHVPEQAFFLIGGLDDLAKKAESLGAKL--------------
MAB_1453|M.abscessus_ATCC_1997 DKLTKGEFDHLPEQAFFLIGGLDDLAKKAESLGAKL--------------
TH_1558|M.thermoresistible__bu DRLTKGDFDHLPEQAFFLIGGLEDLQRKAESLGAKMEDTSGDGAPVPADA
*:* **:***:***********:** :********:
MSMEG_4936|M.smegmatis_MC2_155 ---------
Mvan_4328|M.vanbaalenii_PYR-1 ---------
Mflv_2318|M.gilvum_PYR-GCK ---------
Mb1342|M.bovis_AF2122/97 ---------
Rv1310|M.tuberculosis_H37Rv ---------
MMAR_4087|M.marinum_M ---------
MUL_3954|M.ulcerans_Agy99 ---------
MLBr_01145|M.leprae_Br4923 ---------
MAV_1527|M.avium_104 ---------
MAB_1453|M.abscessus_ATCC_1997 ---------
TH_1558|M.thermoresistible__bu QKDAANTNE