For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPRDEKPVGVAVLGLGNVGSEVVRIIEDSAGDLAARIGAPLVLRGVGVRRVADDRGVPVELLTDDIEGLA ARDDVDIVVELMGPVEPSRKAILCALEHGKSVVTANKALLSTSTGELAQAAESAHVDLYFEAAVAGAIPV IRPLTQSLAGDTVLRVAGIVNGTTNYILSAMDSTGADYDTALAEASDLGYAEADPTADVEGYDAAAKAAI LASIAFHTRVTADDVYREGITKITPADFASARALGCTIKLLSICERITGDDGQERVSARVYPALVPLSHP LATVNGAFNAVVVEARASGRLMFYGQGAGGAPTASAVAGDLVMAARNRVLGSRGPRESKYAQLPVAPMGF ISTRYYVSMNVADKPGVLSSVAAEFAKREVSIAEVRQEGVADEGGRRVGARIVVVTHTATDAALSETVHA LADLDVVQSVTSVLRLEGTSL
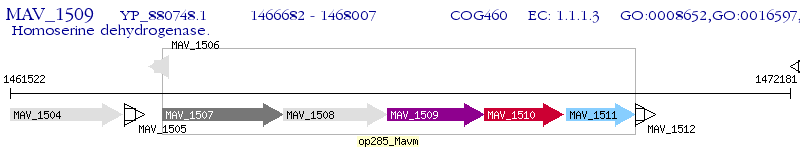
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1509 | - | - | 100% (441) | homoserine dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1326 | thrA | 0.0 | 90.93% (441) | homoserine dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2298 | - | 0.0 | 83.83% (433) | homoserine dehydrogenase |
| M. tuberculosis H37Rv | Rv1294 | thrA | 0.0 | 90.93% (441) | homoserine dehydrogenase |
| M. leprae Br4923 | MLBr_01129 | hom | 0.0 | 89.73% (438) | homoserine dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1435 | - | 0.0 | 81.48% (432) | homoserine dehydrogenase |
| M. marinum M | MMAR_4103 | thrA | 0.0 | 90.25% (441) | homoserine dehydrogenase ThrA |
| M. smegmatis MC2 155 | MSMEG_4957 | - | 0.0 | 85.55% (436) | homoserine dehydrogenase |
| M. thermoresistible (build 8) | TH_2004 | thrA | 0.0 | 87.30% (433) | PROBABLE HOMOSERINE DEHYDROGENASE THRA |
| M. ulcerans Agy99 | MUL_3970 | thrA | 0.0 | 89.57% (441) | homoserine dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4348 | - | 0.0 | 84.76% (433) | homoserine dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2298|M.gilvum_PYR-GCK ---MS---DEIGVAVLGLGNVGSQVVRIIEESATDLTARIGAPLVVRGIG
Mvan_4348|M.vanbaalenii_PYR-1 ---MS---DEIGVAVLGLGNVGSEVVRIIEESAADLTARIGAPLVIRGIG
MSMEG_4957|M.smegmatis_MC2_155 ---MSK--KPIGVAVLGLGNVGSEVVRIIADSADDLAARIGAPLELRGVG
Mb1326|M.bovis_AF2122/97 ---MPGDEKPVGVAVLGLGNVGSEVVRIIENSAEDLAARVGAPLVLRGIG
Rv1294|M.tuberculosis_H37Rv ---MPGDEKPVGVAVLGLGNVGSEVVRIIENSAEDLAARVGAPLVLRGIG
MMAR_4103|M.marinum_M ---MSGDVKPVGVAVLGLGNVGSEVVRIIEESADDLAARVGAPLVLRGVG
MUL_3970|M.ulcerans_Agy99 ---MSGDVKPVGVAVLGLGNVGSEVVRIIEESADDLAARVGAPLVLRGVG
MAV_1509|M.avium_104 ---MPRDEKPVGVAVLGLGNVGSEVVRIIEDSAGDLAARIGAPLVLRGVG
MLBr_01129|M.leprae_Br4923 ---MFSDERTVGVAVLGLGNVGSEVVRIIEGSADDLAARIGAPLMLRGIG
TH_2004|M.thermoresistible__bu -VTMSTRRKPVGVAVLGLGNVGSEVVRIINESATDLAARIGAPLELRGIG
MAB_1435|M.abscessus_ATCC_1997 MS------DAIGVAILGLGNVGSEVARIIESSASDLAARIGAPLEIRGIG
:***:********:*.*** ** **:**:**** :**:*
Mflv_2298|M.gilvum_PYR-GCK VRDVSRDRGVPAELLTDDIDALVSRDDVDIVVELMGPVEPARKAILAALE
Mvan_4348|M.vanbaalenii_PYR-1 VRRVDADRGVPVELLTDDIEALVSRDDVDIVVELMGPVEPARKAILAALE
MSMEG_4957|M.smegmatis_MC2_155 VRRVADDRGVPTELLTDDIDALVSRDDVDIVVEVMGPVEPARKAILSALE
Mb1326|M.bovis_AF2122/97 VRRVTTDRGVPIELLTDDIEELVAREDVDIVVEVMGPVEPSRKAILGALE
Rv1294|M.tuberculosis_H37Rv VRRVTTDRGVPIELLTDDIEELVAREDVDIVVEVMGPVEPSRKAILGALE
MMAR_4103|M.marinum_M VRRVADDRGVPIDLLTDDIEKLVSREDVDIVVEVMGPVEPSRKAILTALE
MUL_3970|M.ulcerans_Agy99 VRRVADDRGVPIDLLTDDIEKLVSREDVDIVVEVMGPVEPSRKAILTALE
MAV_1509|M.avium_104 VRRVADDRGVPVELLTDDIEGLAARDDVDIVVELMGPVEPSRKAILCALE
MLBr_01129|M.leprae_Br4923 VRRVAVDRGVPVDLLTDNIEELVSRADVDIVVEVMGPVELSRKAILSALE
TH_2004|M.thermoresistible__bu VRRVSADRGVPVELLTDNIEELVSRDDVDIVVELMGPVEPARKAILTALE
MAB_1435|M.abscessus_ATCC_1997 VRRVADGRGVPVDLLTDDIEALVARDDVDIVVELMGPVEPARRGILGALN
** * .**** :****:*: *.:* *******:***** :*:.** **:
Mflv_2298|M.gilvum_PYR-GCK QGKSVVTANKALMAVSAGKLAEAAENARVDLYFEAAVAGAIPVIRPLTQS
Mvan_4348|M.vanbaalenii_PYR-1 QGKSVVTANKALMAVSAGKLAEAAEKAHVDLYFEAAVAGAIPVIRPLTQS
MSMEG_4957|M.smegmatis_MC2_155 QGKSVVTANKALMAMSTGELAQAAEKAHVDLYFEAAVAGAIPVIRPLTQS
Mb1326|M.bovis_AF2122/97 RGKSVVTANKALLATSTGELAQAAESAHVDLYFEAAVAGAIPVIRPLTQS
Rv1294|M.tuberculosis_H37Rv RGKSVVTANKALLATSTGELAQAAESAHVDLYFEAAVAGAIPVIRPLTQS
MMAR_4103|M.marinum_M HGKSVVTANKALLSTSTGELAQAAESAHVDLYFEAAVAGAIPVIRPLTQS
MUL_3970|M.ulcerans_Agy99 HGKSVVTANKALLSTSTGELAQAAESAHVDLYFEAAVAGAIPVIRPLTQS
MAV_1509|M.avium_104 HGKSVVTANKALLSTSTGELAQAAESAHVDLYFEAAVAGAIPVIRPLTQS
MLBr_01129|M.leprae_Br4923 HGKSVVTANKALLAASTGELAQAAESAHVDLYFEAAVAGAIPVIRPLTQS
TH_2004|M.thermoresistible__bu QGKSVVTANKALMAQSTGELAQAAENAHVDLYFEASVAGAIPVIRPLTQS
MAB_1435|M.abscessus_ATCC_1997 AGKSVVTANKALLAQSAGELAEAAEKARVDLYFEAAVAGAIPVIRPLTQS
***********:: *:*:**:***.*:*******:**************
Mflv_2298|M.gilvum_PYR-GCK LAGDTVLRVAGIVNGTTNYILSEMDSTGADYASALADASALGYAEADPTA
Mvan_4348|M.vanbaalenii_PYR-1 LAGDTVLRVAGIVNGTTNYILSEMDSTGADYASALADASALGYAEADPTA
MSMEG_4957|M.smegmatis_MC2_155 LAGDTVRRVAGIVNGTTNYILSEMDSTGADYTSALADASALGYAEADPTA
Mb1326|M.bovis_AF2122/97 LAGDTVLRVAGIVNGTTNYILSAMDSTGADYASALADASALGYAEADPTA
Rv1294|M.tuberculosis_H37Rv LAGDTVLRVAGIVNGTTNYILSAMDSTGADYASALADASALGYAEADPTA
MMAR_4103|M.marinum_M LAGDTVLRVAGIVNGTTNYILSEMDSTGADYAKALADAGALGYAEADPTA
MUL_3970|M.ulcerans_Agy99 LAGDTVLRVAGIVNGTTNYILSEMDSTGADYAKALADAGALGYAEADPTA
MAV_1509|M.avium_104 LAGDTVLRVAGIVNGTTNYILSAMDSTGADYDTALAEASDLGYAEADPTA
MLBr_01129|M.leprae_Br4923 LAGDTVLRVAGIVNGTTNYILSAMDSTGADYDSALAGARALGYAEADPTA
TH_2004|M.thermoresistible__bu LAGDTVQRVAGIVNGTTNYILSEMASTGADYDTALAEASALGYAEADPTA
MAB_1435|M.abscessus_ATCC_1997 LAGDSVIRVSGIVNGTTNYILSAMDNTGAGYGTALAEAGELGYAEADPTA
****:* **:************ * .***.* .*** * **********
Mflv_2298|M.gilvum_PYR-GCK DVEGYDAAAKAAILASIAFHTRVTADDVYREGITKVTAADFVSARALGCT
Mvan_4348|M.vanbaalenii_PYR-1 DVEGWDAAAKAAILASIAFHTRVTADDVYREGITKVSAADFASARALGCT
MSMEG_4957|M.smegmatis_MC2_155 DVEGYDAAAKAAILASIAFHTRVTADDVYREGITTVSAEDFASARALGCT
Mb1326|M.bovis_AF2122/97 DVEGYDAAAKAAILASIAFHTRVTADDVYREGITKVTPADFGSAHALGCT
Rv1294|M.tuberculosis_H37Rv DVEGYDAAAKAAILASIAFHTRVTADDVYREGITKVTPADFGSAHALGCT
MMAR_4103|M.marinum_M DVEGFDAAAKAAILASIAFHTRVTADDVYREGITKITPADFASARALGCT
MUL_3970|M.ulcerans_Agy99 DVEGFDAAAKAAILASIAFHTRVTADDVYREGITKITPADFASARALGCT
MAV_1509|M.avium_104 DVEGYDAAAKAAILASIAFHTRVTADDVYREGITKITPADFASARALGCT
MLBr_01129|M.leprae_Br4923 DVEGHDAAAKAAILASIAFHTRVTADDVYREGITKITPADFVSARALGCT
TH_2004|M.thermoresistible__bu DVEGYDAAAKAAILASIAFHTRVTADDVYREGITKVTPADFESARALGCT
MAB_1435|M.abscessus_ATCC_1997 DVEGYDAAAKAAILASIAFHTKVSADDVYREGITKISADDFDSARALQCS
**** ****************:*:**********.::. ** **:** *:
Mflv_2298|M.gilvum_PYR-GCK IKLLAICERLTSPKGKQKVSARVYPALVPLEHPLASVNGAFNAVVVEAEA
Mvan_4348|M.vanbaalenii_PYR-1 IKLLAICERLTTGKGKQQVSARVYPALVPLEHPLASVNGAFNAVVVEAEA
MSMEG_4957|M.smegmatis_MC2_155 IKLLAICERLTSDEGKDRVSARVYPALVPLTHPLAAVNGAFNAVVVEAEA
Mb1326|M.bovis_AF2122/97 IKLLSICERITTDEGSQRVSARVYPALVPLSHPLAAVNGAFNAVVVEAEA
Rv1294|M.tuberculosis_H37Rv IKLLSICERITTDEGSQRVSARVYPALVPLSHPLAAVNGAFNAVVVEAEA
MMAR_4103|M.marinum_M IKLLSICERLTTDEGQQRVSARVYPALVPLSHPLATVNGAFNAVVVEAEA
MUL_3970|M.ulcerans_Agy99 IKLLSICERLTTDEGQQRVSARVYPALVPLSHPLATVNGAFNAVVVEAEA
MAV_1509|M.avium_104 IKLLSICERITGDDGQERVSARVYPALVPLSHPLATVNGAFNAVVVEARA
MLBr_01129|M.leprae_Br4923 IKLLFICERITAADGQQRVSARVYPALVPMSHPLATVSGAFNAVVVEAEA
TH_2004|M.thermoresistible__bu IKLLAICERFTGDDGQERVSARVYPALVPLDHPLAAVNGAFNAVVVEAEA
MAB_1435|M.abscessus_ATCC_1997 IKLLSICERLTDSEGHQRVSARVYPALVPLTHPLASVNGAFNAVFVEAEA
**** ****:* .* ::***********: ****:*.******.***.*
Mflv_2298|M.gilvum_PYR-GCK AGRLMFYGQGAGGAPTASAVLGDLVMAARNRVQGGRGPRESKYAKLPVAP
Mvan_4348|M.vanbaalenii_PYR-1 AGRLMFYGQGAGGAPTASAVLGDLVMAARNRVQGGRGPRESKYAKLPVAP
MSMEG_4957|M.smegmatis_MC2_155 AGRLMFYGQGAGGAPTASAVMGDVVMAARNRVQGGRGPRESKYAKLPIAP
Mb1326|M.bovis_AF2122/97 AGRLMFYGQGAGGAPTASAVTGDLVMAARNRVLGSRGPRESKYAQLPVAP
Rv1294|M.tuberculosis_H37Rv AGRLMFYGQGAGGAPTASAVTGDLVMAARNRVLGSRGPRESKYAQLPVAP
MMAR_4103|M.marinum_M AGRLMFYGQGAGGAPTASAVTGDLVMAARNRVLGSRGPRESKYAQLPVAP
MUL_3970|M.ulcerans_Agy99 AGRLMFYGQGAGGAPTASAVTGDLVMAARNRVLGSRGPRESKYAQLPVAP
MAV_1509|M.avium_104 SGRLMFYGQGAGGAPTASAVAGDLVMAARNRVLGSRGPRESKYAQLPVAP
MLBr_01129|M.leprae_Br4923 AGRLMFYGQGAGGAPTASAVTGDLVMAARNRVLGSRGPKESKYAQLPMET
TH_2004|M.thermoresistible__bu AGRLMFYGQGAGGAPTASAVLGDLVMAARNRVLGGRGPRESKYAKLPVAP
MAB_1435|M.abscessus_ATCC_1997 AGELMFYGQGAGGAPTASAVLGDLVMAARNRVQGGRGPRASRYAQLPVAP
:*.***************** **:******** *.***: *:**:**: .
Mflv_2298|M.gilvum_PYR-GCK IGVIPTRYYVNMNVKDRPGVLSAVAAEFSAREVSIAEVRQEGMVDEDGQR
Mvan_4348|M.vanbaalenii_PYR-1 IGVIPTRYYVNMNVADRPGVLSAVSAEFSKREVSIAEVRQEGMVDDEGQR
MSMEG_4957|M.smegmatis_MC2_155 IGFIPTRYYVNMNVADRPGVLSAVAAEFAKREVSISEVRQEGMVDEVGKP
Mb1326|M.bovis_AF2122/97 MGFIETRYYVSMNVADKPGVLSAVAAEFAKREVSIAEVRQEGVVDEGGRR
Rv1294|M.tuberculosis_H37Rv MGFIETRYYVSMNVADKPGVLSAVAAEFAKREVSIAEVRQEGVVDEGGRR
MMAR_4103|M.marinum_M MGFISTRYYVSMNVADKAGVLSAVAAEFAKRDVSIAEVRQEGVVDQDGQR
MUL_3970|M.ulcerans_Agy99 MGFISTRYYVSINVADKAGVLSAVAAEFAKRDVSIAEVRQKGVVDQDGQR
MAV_1509|M.avium_104 MGFISTRYYVSMNVADKPGVLSSVAAEFAKREVSIAEVRQEGVADEGGRR
MLBr_01129|M.leprae_Br4923 IGFISTRYYVSMNVADKPGVLSGVAAEFAKREVSIAEVRQEGVVDDGGRR
TH_2004|M.thermoresistible__bu IGVIATRYYVSLFVSDKPGVLASVAAEFAKREVSIAEVRQEGVIDEGGER
MAB_1435|M.abscessus_ATCC_1997 IGLIPTRYYVRMRVADKPGVLATVAAEFAKREVSIATVRQEGEVDETG--
:*.* ***** : * *:.***: *:***: *:***: ***:* *: *
Mflv_2298|M.gilvum_PYR-GCK SGARIVVVTHEATDEALSETVAALADLDVVQSINSVLRLEGTTE
Mvan_4348|M.vanbaalenii_PYR-1 CGARIVVVTHQAKDAALSETVAALADLDVVQSINSVLRMEGTSE
MSMEG_4957|M.smegmatis_MC2_155 CGARIVVVTHSATDAALSETVEALADLDVVQNINSVLRLEGTNS
Mb1326|M.bovis_AF2122/97 VGARIVVVTHLATDAALSETVDALDDLDVVQGVSSVIRLEGTGL
Rv1294|M.tuberculosis_H37Rv VGARIVVVTHLATDAALSETVDALDDLDVVQGVSSVIRLEGTGL
MMAR_4103|M.marinum_M IGARIVVVTHLATDAALSETVEALADLDVVENVASVLRLEGNDL
MUL_3970|M.ulcerans_Agy99 IGARIVVVTHLATDAALSETVEVLADLDVVENVASVLRLEGNDL
MAV_1509|M.avium_104 VGARIVVVTHTATDAALSETVHALADLDVVQSVTSVLRLEGTSL
MLBr_01129|M.leprae_Br4923 VGARIVVVTHGATDAALSETVDALADLDVVQGVTSVLRLEGISL
TH_2004|M.thermoresistible__bu IGARIAVLTHSATDAALSETIEALGDLDSVYNVASVLRMEGAGD
MAB_1435|M.abscessus_ATCC_1997 --ARLVVLTHKATDAALSETVAALADLDVVLGVASVIRLEGTQE
**:.*:** *.* *****: .* *** * .: **:*:**