For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LCIPAVVAAALTVGSGIAVADDDGYLAQLKKIGVAWQPGGDTTLIQLGHAICSDRAAGKTPDELAADVHS GLNSSFNYADATAIVSAAESNYCP*
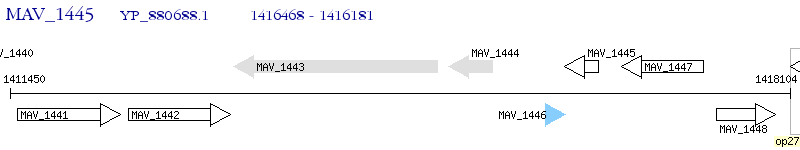
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1445 | - | - | 100% (95) | hypothetical protein MAV_1445 |
| M. avium 104 | MAV_2713 | - | 3e-14 | 40.22% (92) | hypothetical protein MAV_2713 |
| M. avium 104 | MAV_2912 | - | 2e-09 | 35.71% (98) | hypothetical protein MAV_2912 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1302c | - | 1e-05 | 32.22% (90) | hypothetical protein Mb1302c |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv1271c | - | 1e-05 | 32.22% (90) | hypothetical protein Rv1271c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1773c | - | 6e-07 | 36.08% (97) | hypothetical protein MAB_1773c |
| M. marinum M | MMAR_2929 | - | 2e-22 | 48.39% (93) | hypothetical protein MMAR_2929 |
| M. smegmatis MC2 155 | MSMEG_5766 | - | 5e-06 | 34.65% (101) | hypothetical protein MSMEG_5766 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4013 | - | 2e-07 | 30.00% (90) | hypothetical protein MUL_4013 |
| M. vanbaalenii PYR-1 | Mvan_5103 | - | 2e-05 | 29.03% (93) | hypothetical protein Mvan_5103 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1445|M.avium_104 ------------MLCIPAVVAAALTVGSG-IAVAD--DDGYLAQLKKIGV
MMAR_2929|M.marinum_M ----MSARHWLAACTAPVAVGAALAFGTA-VATADATDDAFLAQMHKLGF
Mb1302c|M.bovis_AF2122/97 MLSPLSPRIIAAFTTAVGAAAIGLAVATAGTAGANTKDEAFIAQMESIGV
Rv1271c|M.tuberculosis_H37Rv MLSPLSPRIIAAFTTAVGAAAIGLAVATAGTAGANTKDEAFIAQMESIGV
MUL_4013|M.ulcerans_Agy99 MFS---TRITAAVTTAIGAAAVGLAIATAGSAAASTDDTAFISQMESVGV
MSMEG_5766|M.smegmatis_MC2_155 ----MIFRRQTAWAVAAAAIASGVAFGTA-PAHASVEDDRYLKIVEQLDI
Mvan_5103|M.vanbaalenii_PYR-1 --------MKITIACAASAAFAAVALIGAPAALADP-EGDFLTVIGNGGI
MAB_1773c|M.abscessus_ATCC_199 ----------MIKIAAAAAVAASIVFAPA--AYAD--DDAYLDELSGQGF
. .:.. . * *. : :: : ..
MAV_1445|M.avium_104 A--WQPGGDTTLIQLGHAICSDRAAGKTP--DELAADVHSGLN-SSFNYA
MMAR_2929|M.marinum_M T--WPAGDDSDIVSMGHQICADRMSGKTP--DAIAQDIHTTLGEKGITFA
Mb1302c|M.bovis_AF2122/97 T--FSS--PQVATQQAQLVCKKLASGETG--TEIAEEVLSQTN---LTTK
Rv1271c|M.tuberculosis_H37Rv T--FSS--PQVATQQAQLVCKKLASGETG--TEIAEEVLSQTN---LTTK
MUL_4013|M.ulcerans_Agy99 T--FSS--PQAADREGHQVCQELASGKTG--TDIAEEILSQTD---LTSK
MSMEG_5766|M.smegmatis_MC2_155 P--GEP---EAAIQVGHEICKAVEAGKIEPARTVRGVINHLMNQAGISKG
Mvan_5103|M.vanbaalenii_PYR-1 T--WPSDKTQQVIETGYAVCQDWDNGASF--EQEVADLTSVTD---WSDY
MAB_1773c|M.abscessus_ATCC_199 QVMWQS--RPFLLAAGNGMCNDLRNGETP--EQVASHSNYPNA----TPA
. . :* * .
MAV_1445|M.avium_104 DATAIVSAAESNYCP--------
MMAR_2929|M.marinum_M DVTSMVSAAESTYCSD-------
Mb1302c|M.bovis_AF2122/97 QAAYFVVDATKAYCPQYASQLT-
Rv1271c|M.tuberculosis_H37Rv QAAYFVVDATKAYCPQYASQLT-
MUL_4013|M.ulcerans_Agy99 QAAYFVVYATKDYCPQYASQLA-
MSMEG_5766|M.smegmatis_MC2_155 QAAGLVRGAVSVYCPQYTSLVGR
Mvan_5103|M.vanbaalenii_PYR-1 QAGYFIGAATGAFCPQYEWKVS-
MAB_1773c|M.abscessus_ATCC_199 NLLAMARSAKRNLCP--------
: : * *.