For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRFLARRLLNYLVLLALASFLTFCLTSVAFKPLDSLLQRSPRPPQAVIDAKAHSLGLDEPIPIRYAHWAS HAVRGDFGKTVTGQPVGASLGRRVGVTLRLLVIGSLIGTVAGIAAGAWGAIRQYRLSDRVVTMLALLVLS TPTFVIASLLILAALRVNWALGVQVFDYTGETSPGVTGGAAALLDRLRHLVLPSLTLALAAAAGYSRYQR NAMLDVLGQDFIRTARAKGLTRRRALVKHGLRTALIPLATLFAYGVAGLVTGAVFVEKIFGWHGMGEWLV QGVATQDTNIVAAITLFSGAVVLLAGLLSDVFYAALDPRVRVS*
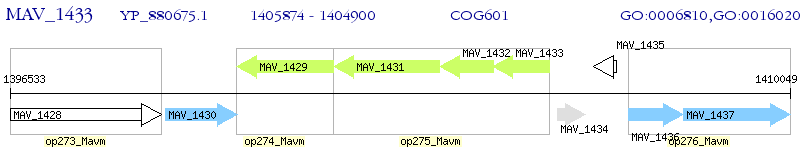
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1433 | oppB | - | 100% (324) | ABC transporter, permease protein OppB |
| M. avium 104 | MAV_0465 | dppB | 3e-34 | 30.43% (322) | ABC transporter, permease protein DppB |
| M. avium 104 | MAV_3420 | - | 8e-19 | 26.25% (320) | ABC transporter permease protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1314c | oppB | 1e-158 | 84.92% (325) | oligopeptide-transport integral membrane protein ABC |
| M. gilvum PYR-GCK | Mflv_2261 | - | 1e-138 | 75.69% (325) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv1283c | oppB | 1e-158 | 84.92% (325) | oligopeptide-transport integral membrane protein ABC |
| M. leprae Br4923 | MLBr_01124 | - | 1e-159 | 84.62% (325) | putative binding-protein dependent transport protein |
| M. abscessus ATCC 19977 | MAB_0427 | - | 2e-37 | 30.43% (322) | peptide ABC transporter transmembrane protein |
| M. marinum M | MMAR_4136 | oppB | 1e-159 | 84.92% (325) | oligopeptide-transport integral membrane protein ABC |
| M. smegmatis MC2 155 | MSMEG_4995 | - | 1e-144 | 78.15% (325) | oligopeptide transport system permease protein AppB |
| M. thermoresistible (build 8) | TH_4246 | - | 1e-146 | 80.50% (323) | PUTATIVE oligopeptide transport system permease protein AppB |
| M. ulcerans Agy99 | MUL_3999 | oppB | 1e-158 | 84.31% (325) | oligopeptide-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_4430 | - | 1e-139 | 76.62% (325) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2261|M.gilvum_PYR-GCK ----------------MTRFLVRRLLKFVILLGVASFLTFALT-SVMFQP
Mvan_4430|M.vanbaalenii_PYR-1 ----------------MTRFLARRLLNYVVLLGLASFLTFTLT-SMTFSP
MSMEG_4995|M.smegmatis_MC2_155 ----------------MTRFLARRLLNYVILLALASVLTFTLT-SLSFDP
TH_4246|M.thermoresistible__bu ----------------VTRFLARRFLNYLILLALAAFLTFTLT-SVSFRP
Mb1314c|M.bovis_AF2122/97 ----------------MTRYLARRLLNYLVLLALASFLTYCLT-SLAFSP
Rv1283c|M.tuberculosis_H37Rv ----------------MTRYLARRLLNYLVLLALASFLTYCLT-SLAFSP
MMAR_4136|M.marinum_M ----------------MTRFLARRLLNYVVLLALASFLTYCLT-SLAFSP
MUL_3999|M.ulcerans_Agy99 ----------------MTRFLARRLLNYVVLLALASFLTYCLT-SLAFSP
MAV_1433|M.avium_104 ----------------MTRFLARRLLNYLVLLALASFLTFCLT-SVAFKP
MLBr_01124|M.leprae_Br4923 ----------------MMRFLARRLLNYVVLLALASFLTYCLT-SMAFAP
MAB_0427|M.abscessus_ATCC_1997 MTTVLGNVRQFWQSAGLFRYIAKRLLLAIPVLIGTSLLIYSLVYALPGDP
: *::.:*:* : :* ::.* : *. :: *
Mflv_2261|M.gilvum_PYR-GCK LDSLLERNPRPPQAVIDAKAEELNLNAPIPVRYADWVSGAVRGDFGTTVG
Mvan_4430|M.vanbaalenii_PYR-1 LDSLLERNPRPPQAVIDAKAAELNLDKPIPVRYGDWVAGAVRGDFGTTVG
MSMEG_4995|M.smegmatis_MC2_155 LDSLLQRNPRPPQSTIDAKAAELGLDKPIPLRYLHWASGAVRGDFGTTVT
TH_4246|M.thermoresistible__bu LDSLEQRNPRPPQAVIDAKAAELGLDKPIPIRFADWAGGAVRGDFGTTIT
Mb1314c|M.bovis_AF2122/97 LESLMQRSPRPPQAVIDAKAHDLGLDRPILARYANWVSHAVRGDFGTTIT
Rv1283c|M.tuberculosis_H37Rv LESLMQRSPRPPQAVIDAKAHDLGLDRPILARYANWVSHAVRGDFGTTIT
MMAR_4136|M.marinum_M LESLMQRSPRPPQAVIDAKAAELGLNKPIPVRYAHWVTHAVRGDFGVTIS
MUL_3999|M.ulcerans_Agy99 LESLMQRSPRPPQAVIDAKAAELGLNKPIPVRYAHWVTHAVRGDFGVTIS
MAV_1433|M.avium_104 LDSLLQRSPRPPQAVIDAKAHSLGLDEPIPIRYAHWASHAVRGDFGKTVT
MLBr_01124|M.leprae_Br4923 LDSLLQRSPHPPQAMVDAKAHALGLDKPIPIRYANWASHAIRGDFGTTIT
MAB_0427|M.abscessus_ATCC_1997 IRALAGDRPFTP-EVMAQLRERFHLNDPFLVRYGKYIVGFLHGDFGTDFQ
: :* * .* : : *: *: *: .: ::**** .
Mflv_2261|M.gilvum_PYR-GCK GQPVSDELWRRIGVSLRLVLIGSVIGTVLGVVAGAWGAIRQYRLSDRVIT
Mvan_4430|M.vanbaalenii_PYR-1 GQPVTDELWRRIGVSLRLVIIGSIVGTFLGVVAGAWGAVRQYRLSDRVIT
MSMEG_4995|M.smegmatis_MC2_155 GQPVSEELWRRIGVSLRLLLLGSILGTVIGVVVGAWGAIRQYRLSDRVVT
TH_4246|M.thermoresistible__bu GQPVSEELWRRIGVSLRLLIIGSVLGTVAGVVLGAWGAIRQYRLSDRVVT
Mb1314c|M.bovis_AF2122/97 GQPVGTELGRRIGVSLRLLVVGSVFGTVAGVVIGAWGAIRQYRLSDRVMT
Rv1283c|M.tuberculosis_H37Rv GQPVGTELGRRIGVSLRLLVVGSVFGTVAGVVIGAWGAIRQYRLSDRVMT
MMAR_4136|M.marinum_M GQPVSAELGRRIGTSLRLLLIGSVVGTALGVAIGAWGAIRQYRFSDRVIT
MUL_3999|M.ulcerans_Agy99 GQPVSAELGRRIGTSLRLLLIGSVVGAALGVAIGAWGAIRQYRFSDRVIT
MAV_1433|M.avium_104 GQPVGASLGRRVGVTLRLLVIGSLIGTVAGIAAGAWGAIRQYRLSDRVVT
MLBr_01124|M.leprae_Br4923 GQPVGSTLGRRVGVSLRLLIIGSLTGTVLGVTAGAWGAIRQYQLSDRVVT
MAB_0427|M.abscessus_ATCC_1997 QRPVAQTVSQRLPVTLRLTVVAVAFETIIGITAGVLCAVRRGSFYDNLVL
:** : :*: .:*** ::. : *:. *. *:*: : *.::
Mflv_2261|M.gilvum_PYR-GCK ALALLILSAPTFVIANLLILGALQANSALGVQLFEYTGESSPDAVGGGWN
Mvan_4430|M.vanbaalenii_PYR-1 TLALLTLSAPTFVIANLLILAALNVNSFLGLQVFEYTGETSSDAVGGGWN
MSMEG_4995|M.smegmatis_MC2_155 VVSLLILSTPTFVIANLLILGALDVNSVLGVRLFEYTGETSPDAVGGSWA
TH_4246|M.thermoresistible__bu VLSLLVLSTPTFVIANLLILGALNVNSALGVQLFAYTGETSPDVVGGTWN
Mb1314c|M.bovis_AF2122/97 TLALLVLSTPTFVVANLLILGALRVNWAVGIQLFDYTGETSPGVAGGVWD
Rv1283c|M.tuberculosis_H37Rv TLALLVLSTPTFVVANLLILGALRVNWAVGIQLFDYTGETSPGVAGGVWD
MMAR_4136|M.marinum_M MLALLVLSTPTFVVANLLILGALRVNWALGFQLFDYTGETSPGVDGGTWA
MUL_3999|M.ulcerans_Agy99 MLALLVLSTPTFVVANLLILGALRVNWALGFQLFDYTGETSPGVDGGTWA
MAV_1433|M.avium_104 MLALLVLSTPTFVIASLLILAALRVNWALGVQVFDYTGETSPGVTGGA-A
MLBr_01124|M.leprae_Br4923 LLALLVLSTPTFVIANLLILSALRVNWAFGIQLFDYTGETSPGVYGGAWA
MAB_0427|M.abscessus_ATCC_1997 VTTTLLVSMPVFVLG-------FLAQYLFGFKLGWFP-----------IA
: * :* *.**:. : .: .*.:: :.
Mflv_2261|M.gilvum_PYR-GCK QFVDRAQHLLLPTFTLALTAMAGYSRYQRNAMLDVLGQDFIRTARAKGLT
Mvan_4430|M.vanbaalenii_PYR-1 QFVDRLQHLVLPTFTLALGAIAGFSRYQRNAMLDVLSQDFIRTARAKGLT
MSMEG_4995|M.smegmatis_MC2_155 QFVDRLQHLVLPTLTLALGGIAGFSRYQRNAMLDVLSQDYIRTARAKGLT
TH_4246|M.thermoresistible__bu QILDRLQHLVLPTLTLALAAMAGYSRYQRNAMLDVLGQDFIRTARAKGLT
Mb1314c|M.bovis_AF2122/97 RLGDRLQHLILPSLTLALAAAAGFSRYQRNAMLDVLGQDFIRTARAKGLT
Rv1283c|M.tuberculosis_H37Rv RLGDRLQHLILPSLTLALAAAAGFSRYQRNAMLDVLGQDFIRTARAKGLT
MMAR_4136|M.marinum_M AFGDRLKHLVLPSLTLALGAAAGYSRYQRNAMLDVLGQDFIRTARAKGLT
MUL_3999|M.ulcerans_Agy99 AFGDRLKHLVLPSLTLALGAAAGYSRYQRNAMLDVLGQDFIRTARAKGLT
MAV_1433|M.avium_104 ALLDRLRHLVLPSLTLALAAAAGYSRYQRNAMLDVLGQDFIRTARAKGLT
MLBr_01124|M.leprae_Br4923 HVIDRLRHLILPTLTLALVGAAGYSRYQRNAMLDVLSQDFIRAARAKGLT
MAB_0427|M.abscessus_ATCC_1997 GIRDGWYSFLLPGLVLASLSMAYVARLTRTSMLETMDAEFVRTARAKGIS
. * ::** :.** . * :* *.:**:.:. :::*:*****::
Mflv_2261|M.gilvum_PYR-GCK RRQALFKHGLRTALIPMATLFAYSVGGLFTGAVFVEKIFGWHGVGEWFVL
Mvan_4430|M.vanbaalenii_PYR-1 RRRALFKHGLRTALIPMATLFAYGVGGLFTGAVFVEKIFGWHGIGEWFVQ
MSMEG_4995|M.smegmatis_MC2_155 RRRALFKHGLRTALIPMATLFAYTVAGLVTGAVFVEKIFGWHGMGEWVVQ
TH_4246|M.thermoresistible__bu RRQALVKHGLRTALIPMATLFAYGLAGLVTGSVFVEKIFGWHGMGEWIVF
Mb1314c|M.bovis_AF2122/97 RRRALLKHGLRTALIPMATLFAYGVAGLVTGAVFVEKIFGWHGMGEWMVR
Rv1283c|M.tuberculosis_H37Rv RRRALLKHGLRTALIPMATLFAYGVAGLVTGAVFVEKIFGWHGMGEWMVR
MMAR_4136|M.marinum_M RRRALIKHGLRTALIPMATLFAYGVAGLVTGAVFVEKIFGWHGMGEWAVR
MUL_3999|M.ulcerans_Agy99 RRRALIKHGLRTALIPMATLFAYGVAGLVTGAVFVEKIFGWHGMGEWAVR
MAV_1433|M.avium_104 RRRALVKHGLRTALIPLATLFAYGVAGLVTGAVFVEKIFGWHGMGEWLVQ
MLBr_01124|M.leprae_Br4923 RRRALLKHGLRTALIPMATLFAYGVAGLVTGALFVEKIFGWHGMGEWMVQ
MAB_0427|M.abscessus_ATCC_1997 ESRILLRHTLRNSLIPVVTFIGADVGALLAGAVVTESVFNIPGLGRATFD
. : *.:* **.:***:.*::. :..*.:*::..*.:*. *:*. .
Mflv_2261|M.gilvum_PYR-GCK GIAAQDTNIIVAITVFTGATVLLAGLLSDVLYAALDPRVRVS
Mvan_4430|M.vanbaalenii_PYR-1 GIGSQDTNIVVAITVFTGVTVLLAGLLSDVLYAALDPRVRVS
MSMEG_4995|M.smegmatis_MC2_155 GIATQDTNIIAAITVFTGTVVLLAGLLSDVIYAMLDPRVRVT
TH_4246|M.thermoresistible__bu GIATQDTNIVVAITVFTGAMILVAGLLSDVIYAMLDPRVRAR
Mb1314c|M.bovis_AF2122/97 GISTQDTNIVAAITVFSGAVVLLAGLLSDVIYAALDPRVRVS
Rv1283c|M.tuberculosis_H37Rv GISTQDTNIVAAITVFSGAVVLLAGLLSDVIYAALDPRVRVS
MMAR_4136|M.marinum_M GIATQDTNIVAAITVFSGAVVLLAGLLSDIFYAALDPRVRVS
MUL_3999|M.ulcerans_Agy99 GIATQDTNIVAAITVFSGAAVLLAGLLSDIFYAALDPRVRVS
MAV_1433|M.avium_104 GVATQDTNIVAAITLFSGAVVLLAGLLSDVFYAALDPRVRVS
MLBr_01124|M.leprae_Br4923 GVATQDTNIIAAITIFFGTVILLAGLLSDVIYAALDPRVRVS
MAB_0427|M.abscessus_ATCC_1997 AVRNQEGAVVVSILTLIVFFYIFFNLVVDILYALLDPRIRYE
.: *: ::.:* : :. .*: *::** ****:*