For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
EIAVQGSLFQHTERRQLGDGAFIEIRAGWLTDADDLLDALLSTVPWREERRQMYDRVVDVPRLVSFHDLT VDEPPHPMLSRLRRRLNDIYAGELGEPFTTVGLCCYRDGSDSVAWHGDTIGRSSSEDTMVAIVSLGATRA FALRRRGGGPSLRLPQAHGDLLVMGGSCQRTWEHAVPKTTAPVGPRVSIQFRPRGVR*
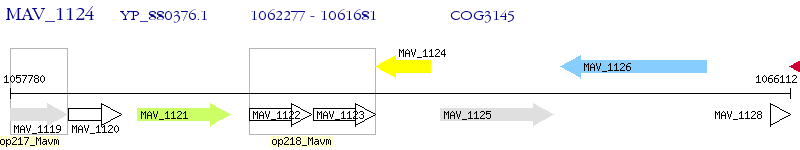
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1124 | - | - | 100% (198) | alkylated DNA repair protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1027c | - | 2e-98 | 84.85% (198) | hypothetical protein Mb1027c |
| M. gilvum PYR-GCK | Mflv_1921 | - | 9e-83 | 71.78% (202) | hypothetical protein Mflv_1921 |
| M. tuberculosis H37Rv | Rv1000c | - | 2e-98 | 84.85% (198) | hypothetical protein Rv1000c |
| M. leprae Br4923 | MLBr_00190 | - | 5e-98 | 83.25% (197) | hypothetical protein MLBr_00190 |
| M. abscessus ATCC 19977 | MAB_1120c | - | 7e-75 | 68.02% (197) | hypothetical protein MAB_1120c |
| M. marinum M | MMAR_4493 | - | 3e-98 | 83.84% (198) | hypothetical protein MMAR_4493 |
| M. smegmatis MC2 155 | MSMEG_5451 | - | 3e-86 | 74.87% (199) | alkylated DNA repair protein |
| M. thermoresistible (build 8) | TH_0932 | - | 2e-81 | 77.47% (182) | CONSERVED HYPOTHETICAL PROTEIN. THOUGHT TO BE REGULATED BY |
| M. ulcerans Agy99 | MUL_4665 | - | 2e-97 | 83.76% (197) | hypothetical protein MUL_4665 |
| M. vanbaalenii PYR-1 | Mvan_4812 | - | 3e-83 | 72.77% (202) | hypothetical protein Mvan_4812 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1921|M.gilvum_PYR-GCK -------MEPALQSSLFEQS--DRRDLGNGAWLEVRSGWLSQDGSSDDTL
Mvan_4812|M.vanbaalenii_PYR-1 -------MEPALQSALFDHS--ERRDLGDGAWLEVRSGWLSQDGAADDAL
MSMEG_5451|M.smegmatis_MC2_155 -------MELAVQGSLFEHA--ERRPLGNGAWIELRSGWLAD----ADSL
TH_0932|M.thermoresistible__bu -------------------------MLGDGAWVEVRCGWLTD----ADTL
Mb1027c|M.bovis_AF2122/97 MCDKLGGVAIAVQGALFEHN--ERRQLGDGAFIDIRSGWLTG----GEEL
Rv1000c|M.tuberculosis_H37Rv MCDKLGGVAIAVQGALFEHN--ERRQLGDGAFIDIRSGWLTG----GEEL
MMAR_4493|M.marinum_M MCDKLGAVAFAVQGSLFEHN--ERRQLGDGAFIDIRANWLRA----GDDL
MUL_4665|M.ulcerans_Agy99 MCDKLGAVAFAVQGSLFEHN--ERRQLGDGAFIDIRANWLRA----GDDL
MAV_1124|M.avium_104 -------MEIAVQGSLFQHT--ERRQLGDGAFIEIRAGWLTD----ADDL
MLBr_00190|M.leprae_Br4923 MCDMLVDVGIAFQGSLFEYH--ERRQLGDGAFIELRSGWLTD----GVEL
MAB_1120c|M.abscessus_ATCC_199 -------MTHAVQASLFGAPPVPRRELAGGAWLEVHHEWLGR----ADNL
*..**::::: ** *
Mflv_1921|M.gilvum_PYR-GCK FAELRDRIPWRAERRQMYDRVLDVPRLVSFHNLLDGNAPHPRLKQLRRRL
Mvan_4812|M.vanbaalenii_PYR-1 FGELRDQIPWRAERRQMYDRVLNVPRLVSFHNLVDAAAPHPRLKQLRRRL
MSMEG_5451|M.smegmatis_MC2_155 FEELMDTIPWRAEQREMYDRVVDVPRLVSFHNLVDEPAPHPRLKQIRRRL
TH_0932|M.thermoresistible__bu FDELVEVIPWRAERRHMYDRMVDVPRLVSFHNLVDEPAPHPRLKQMRRRL
Mb1027c|M.bovis_AF2122/97 LDALLSTVPWRAERRQMYDRVVDVPRLVSFHDLTIEDPPHPQLARMRRRL
Rv1000c|M.tuberculosis_H37Rv LDALLSTVPWRAERRQMYDRVVDVPRLVSFHDLTIEDPPHPQLARMRRRL
MMAR_4493|M.marinum_M LEALISRVPWRSERRQMYDRVVEVPRLVSFHDLMIEKPPHPELARMRRRL
MUL_4665|M.ulcerans_Agy99 LEALISRVPWRSERRQMYDRVVEVPRLVSFHDLMIEKPPHPELARMRRRL
MAV_1124|M.avium_104 LDALLSTVPWREERRQMYDRVVDVPRLVSFHDLTVDEPPHPMLSRLRRRL
MLBr_00190|M.leprae_Br4923 LDTLLSEVPWRIERRRMYDKVVNVPRLVSFHDLTTDDPPHPLLTRLRRRL
MAB_1120c|M.abscessus_ATCC_199 MGQLLAQVPWRAERRRMYDRTLDVPRLVSFHDLTSGPPPHPVLETICDRL
: * :*** *:*.***: ::********:* .*** * : **
Mflv_1921|M.gilvum_PYR-GCK NDVYAGELGEPFVTAGLCLYRDGDDSVAWHGDNIGRSSTEDTMVAIVGIG
Mvan_4812|M.vanbaalenii_PYR-1 NDAYAGELGEPFVTAGLCLYRDGDDSVAWHGDNIGRSSTEDTMVAIVSLG
MSMEG_5451|M.smegmatis_MC2_155 NDTYGRELGEPFTTAGLCLYRDGDDSVAWHGDTIGRSRTRDTMVAIVSLG
TH_0932|M.thermoresistible__bu NDAYAGELGEPFVTAGLAYYRDGSDSVAWHGDTIGRSRTEDTMVAIVSLG
Mb1027c|M.bovis_AF2122/97 NDIYGGELGEPFTTAGLCYYRDGSDSVAWHGDTIGRGSTEDTMVAIVSLG
Rv1000c|M.tuberculosis_H37Rv NDIYGGELGEPFTTAGLCYYRDGSDSVAWHGDTIGRGSTEDTMVAIVSLG
MMAR_4493|M.marinum_M NDIYGGELGEPFTTVGLCCYRDGSDSVAWHGDTIGRSSTEDTMVAIVSLG
MUL_4665|M.ulcerans_Agy99 NDIYGGELGEPFTTVGLCCYRDGFDSVAWHGDTIGRSSTEDTMVAIVSLG
MAV_1124|M.avium_104 NDIYAGELGEPFTTVGLCCYRDGSDSVAWHGDTIGRSSSEDTMVAIVSLG
MLBr_00190|M.leprae_Br4923 NDIYAGELGEPFTSVGLCCYRDGSDSIAWHGDTIGRNSSEDTMVAIISLG
MAB_1120c|M.abscessus_ATCC_199 NGEYTDELGEPFTTVGLCQYRDGTDSVAWHGDTIGRGLREDTMVAIVSIG
*. * ******.:.**. **** **:*****.***. .******:.:*
Mflv_1921|M.gilvum_PYR-GCK ATRVFALRPRGGGPSLRLRHCHGDLLVMGGSCQRTWEHAVPKTTRPTGPR
Mvan_4812|M.vanbaalenii_PYR-1 ATRVFALRPRGGGPSLRIRHCHGDLLVMGGSCQRTWEHAVPKTARPVGPR
MSMEG_5451|M.smegmatis_MC2_155 ATRVFALRPRGGGHALRLQQQHGDLLVMGGSCQRTWEHSVPKTSRLIGPR
TH_0932|M.thermoresistible__bu ATRTLAMRPKGGGRSLRFPLNHGDLLVMGGSCQRTWEHSVPKTTKPTGPR
Mb1027c|M.bovis_AF2122/97 ATRVFALRPRGRGPSLRLPLAHGDLLVMGGSCQRTFEHAVPKTSAPTGPR
Rv1000c|M.tuberculosis_H37Rv ATRVFALRPRGRGPSLRLPLAHGDLLVMGGSCQRTFEHAVPKTSAPTGPR
MMAR_4493|M.marinum_M ATRIFAMRPRGGGASLRLPLAHGDLLVMGGSCQRTWEHSVPKTSTPAGPR
MUL_4665|M.ulcerans_Agy99 ATRIFALRPRGGGASLRLPLAHGDLLVMGGSCQRTWEHSVPKTSTPAGPR
MAV_1124|M.avium_104 ATRAFALRRRGGGPSLRLPQAHGDLLVMGGSCQRTWEHAVPKTTAPVGPR
MLBr_00190|M.leprae_Br4923 ATRVFALRKRGGGPSLRLPLTHGDLLVMGGSCQRTWEHSVPKTSASTGPR
MAB_1120c|M.abscessus_ATCC_199 ATRTFALRPRLGGESIRINIGHGDLLVMGGSCQRTWEHAIPKTSKPVGPR
*** :*:* : * ::*: **************:**::***: ***
Mflv_1921|M.gilvum_PYR-GCK ISIQFRPHDVR
Mvan_4812|M.vanbaalenii_PYR-1 ISIQFRPHDVR
MSMEG_5451|M.smegmatis_MC2_155 ISIQFRPHNVR
TH_0932|M.thermoresistible__bu ISIQFRPRGVR
Mb1027c|M.bovis_AF2122/97 VSIQFRPRDVR
Rv1000c|M.tuberculosis_H37Rv VSIQFRPRDVR
MMAR_4493|M.marinum_M VSIQFRPRGVR
MUL_4665|M.ulcerans_Agy99 VSIQFRPRGVC
MAV_1124|M.avium_104 VSIQFRPRGVR
MLBr_00190|M.leprae_Br4923 VSIQFRPRNVH
MAB_1120c|M.abscessus_ATCC_199 ISLQYRPRDVR
:*:*:**:.*