For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAKRLDLKGVNIYYGSFHAVAEVTLSVLPRSVTAFIGPSGCGKTTVLRTLNRMHEVVPGGRVEGSVLLDD EDIYGPGVDPVGVRRAIGMVFQRPNPFPAMSIRDNVVAGLKLQGVRNRKVLDETAEYSLRGANLWDEVKD RLDRPGGGLSGGQQQRLCIARAIAVQPDVLLMDEPCSALDPISTMAIEELISELKQDYTIVIVTHNMQQA ARVSDYTAFFNLEAVGKPGRLIEVDDTEKIFSNPSQKATEDYISGRFG
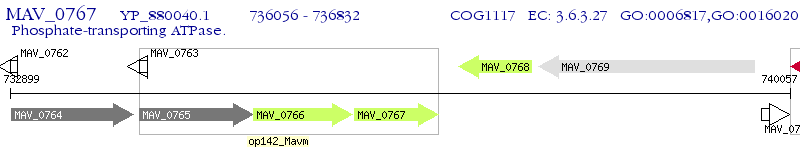
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0767 | pstB | - | 100% (258) | phosphate ABC transporter, ATP-binding protein PstB |
| M. avium 104 | MAV_0668 | pstB | 1e-28 | 41.58% (202) | ABC transporter, ATP-binding protein PstB |
| M. avium 104 | MAV_0318 | - | 4e-28 | 35.86% (237) | amino acid ABC transporter, ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0843 | phoT | 1e-134 | 89.92% (258) | phosphate ABC transporter ATP-binding protein |
| M. gilvum PYR-GCK | Mflv_1664 | - | 1e-131 | 87.98% (258) | phosphate ABC transporter, ATPase subunit |
| M. tuberculosis H37Rv | Rv0820 | phoT | 1e-135 | 90.31% (258) | phosphate ABC transporter ATP-binding protein |
| M. leprae Br4923 | MLBr_02189 | phoT | 1e-136 | 91.09% (258) | phosphate transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_0749 | - | 1e-127 | 85.66% (258) | phosphate ABC transporter ATP-binding protein |
| M. marinum M | MMAR_4860 | phoT | 1e-140 | 94.19% (258) | phosphate-transport ATP-binding protein ABC transporter, |
| M. smegmatis MC2 155 | MSMEG_5779 | pstB | 1e-132 | 88.76% (258) | phosphate ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_2660 | - | 1e-130 | 87.21% (258) | PUTATIVE phosphate ABC transporter, ATP-binding protein |
| M. ulcerans Agy99 | MUL_0440 | phoT | 1e-140 | 94.19% (258) | phosphate-transport ATP-binding protein ABC transporter, |
| M. vanbaalenii PYR-1 | Mvan_5089 | - | 1e-128 | 88.37% (258) | phosphate ABC transporter, ATPase subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5779|M.smegmatis_MC2_155 MAKRLDLKDVNIYYGAFHAVADVSLAVQPRSVTAFIGPSGCGKSTVLRTL
TH_2660|M.thermoresistible__bu MAKRLDLKDVNIYYGGFHAVADINLSVPPRMVTAFIGPSGCGKSTVLRTL
MAB_0749|M.abscessus_ATCC_1997 MAKRLDLKDVNIFYGKFHAVQDVALSVPPRNVTAFIGPSGCGKSTVLRTL
Mflv_1664|M.gilvum_PYR-GCK MAKRLDLKDLNIYYGSFHAVADVGLSVMPRSVTAFIGPSGCGKSTVLRTL
Mvan_5089|M.vanbaalenii_PYR-1 MAKRLDLKDLNIYYGSFHAVADVGLSVMPRSVTAFIGPSGCGKSTVLRTL
MMAR_4860|M.marinum_M MAKRLDLKDVNIYYGSFHAVAEVSLSILPRSVTAFIGPSGCGKTTVLRTL
MUL_0440|M.ulcerans_Agy99 MAKRLDLKDVNIYYGSFHAVAEVSLSILPRSVTAFIGPSGCGKTTVLRTL
MAV_0767|M.avium_104 MAKRLDLKGVNIYYGSFHAVAEVTLSVLPRSVTAFIGPSGCGKTTVLRTL
Mb0843|M.bovis_AF2122/97 MAKRLDLTDVNIYYGSFHAVADVSLAILPRSVTALIGPSGCGKTTVLRTL
Rv0820|M.tuberculosis_H37Rv MAKRLDLTDVNIYYGSFHAVADVSLAILPRSVTAFIGPSGCGKTTVLRTL
MLBr_02189|M.leprae_Br4923 MAKRLDLKGVNIYYGSFQAVLDVSLAVLPRSVTAFIGASGCGKTTVLRTL
*******..:**:** *:** :: *:: ** ***:**.*****:******
MSMEG_5779|M.smegmatis_MC2_155 NRMHEVIPGARVEGSVLLDGEDIYGPGVDPVGVRKTIGMVFQRPNPFPTM
TH_2660|M.thermoresistible__bu NRMHEVIPGARVEGSVLLDGEDIYAPGVDPVSVRKTIGMVFQRPNPFPTM
MAB_0749|M.abscessus_ATCC_1997 NRMHEVTPGARVEGSVLLDGADIYGSAVDPVSVRKTIGMVFQRPNPFPTM
Mflv_1664|M.gilvum_PYR-GCK NRMHEVIPGARVEGSVLLDGEDIYGAGIDPVSVRKTIGMVFQRPNPFPTM
Mvan_5089|M.vanbaalenii_PYR-1 NRMHEVLPGARVEGSVLLDGEDIYASGVDPVSVRKTIGMVFQRPNPFPTM
MMAR_4860|M.marinum_M NRMHEVIPGARVEGSVLLDDENIYGPGIDPVGVRRAIGMVFQRPNPFPAM
MUL_0440|M.ulcerans_Agy99 NRMHEVIPGARVEGSVLLDDENIYGPGIDPVGVRRAIGMVFQRPNPFPAM
MAV_0767|M.avium_104 NRMHEVVPGGRVEGSVLLDDEDIYGPGVDPVGVRRAIGMVFQRPNPFPAM
Mb0843|M.bovis_AF2122/97 NRMHEVIPGARVEGAVLLDDQDIYAPGIDPVGVRRAIGMVFQRPNPFPAM
Rv0820|M.tuberculosis_H37Rv NRMHEVIPGARVEGAVLLDDQDIYAPGIDPVGVRRAIGMVFQRPNPFPAM
MLBr_02189|M.leprae_Br4923 NRMHEVVPGARVEGTVLLDDEDIYAPGIDPVGVRRAIGMVFQRPNPFPAM
****** **.****:****. :**...:***.**::************:*
MSMEG_5779|M.smegmatis_MC2_155 SIRDNVVAGLKLQGVRNKKTLDEVAERSLRGANLWNEVKDRLDKPGGGLS
TH_2660|M.thermoresistible__bu SIRDNVVAGLKLQGVRNKKVLDEVCERSLRGANLWNEVKDRLDKPGGGLS
MAB_0749|M.abscessus_ATCC_1997 SIRDNVVAGLRLQGVRNKKTLDEVAERSLQGANLWNEVKDRLDRPGGGLS
Mflv_1664|M.gilvum_PYR-GCK SIRDNVVAGLKLQGIRNKKTLDEIAVSSLKGANLWNEVKDRLDKPGGGLS
Mvan_5089|M.vanbaalenii_PYR-1 SIRDNVVAGLKLQGVR--RNLDDIAEKSLKGANLWNEVKDRLDKPGGGLS
MMAR_4860|M.marinum_M SIRDNVVAGLKLQGVRNRKVLDETAEYSLRGANLWDEVKDRMDRPGGGLS
MUL_0440|M.ulcerans_Agy99 SIRDNVVAGLKLQGVRNRKVLDETAEYSLRGANLWDEVKDRMDRPGGGLS
MAV_0767|M.avium_104 SIRDNVVAGLKLQGVRNRKVLDETAEYSLRGANLWDEVKDRLDRPGGGLS
Mb0843|M.bovis_AF2122/97 SIRNNVVAGLKLQGVRNRKVLDDTAESSLRGANLWDEVKDRLDKPGGGLS
Rv0820|M.tuberculosis_H37Rv SIRNNVVAGLKLQGVRNRKVLDDTAESSLRGANLWDEVKDRLDKPGGGLS
MLBr_02189|M.leprae_Br4923 SIRDNVVAGLKLQGVRNRKVLDDTAEYFLRGTNLWDEVKDRLDKPGGGLS
***:******:***:* : **: . *:*:***:*****:*:******
MSMEG_5779|M.smegmatis_MC2_155 GGQQQRLCIARAIAVQPDVLLMDEPCSALDPISTLAIEDLIATLKLDYTI
TH_2660|M.thermoresistible__bu GGQQQRLCIARAIAVEPDVLLMDEPCSALDPISTLAIEDLIAKLKQDFTI
MAB_0749|M.abscessus_ATCC_1997 GGQQQRLCIARAIAVQPDVLLMDEPCSALDPISTLAIEDLISELKQEFTI
Mflv_1664|M.gilvum_PYR-GCK GGQQQRLCIARAIAVQPDVLLMDEPCSALDPISTLAIEDLISELKQDYTI
Mvan_5089|M.vanbaalenii_PYR-1 GGQQQRLCIARAIAVQPDVLLMDEPCSALDPISTLAIEDLISELKQDYTI
MMAR_4860|M.marinum_M GGQQQRLCIARAIAVQPDVLLMDEPCSALDPISTMAIEDLISELKQEYTI
MUL_0440|M.ulcerans_Agy99 GGQQQRLCIARAIAVQPDVLLMDEPCSALDPISTMAIEDLISELKQEYTI
MAV_0767|M.avium_104 GGQQQRLCIARAIAVQPDVLLMDEPCSALDPISTMAIEELISELKQDYTI
Mb0843|M.bovis_AF2122/97 GGQQQRLCIARAIAVQPDVLLMDEPCSSLDPISTMAIEDLISELKQQYTI
Rv0820|M.tuberculosis_H37Rv GGQQQRLCIARAIAVQPDVLLMDEPCSSLDPISTMAIEDLISELKQQYTI
MLBr_02189|M.leprae_Br4923 GGQQQRLCIARAIAVQPDVLLMDEPCSSLDPISTMAIEELIGELKQEYTI
***************:***********:******:***:**. ** ::**
MSMEG_5779|M.smegmatis_MC2_155 VIVTHNMQQAARVSDQTAFFNLEATGKPGRLIEIDDTEKIFSNPRQKATE
TH_2660|M.thermoresistible__bu VIVTHNMQQAARVSDQTAFFNLEATGRPGRLIEIDDTEKIFSNPSQKATE
MAB_0749|M.abscessus_ATCC_1997 VIVTHNMQQAARVSDQTAFFNLEAAGKPGMLVEIDDTEKIFSNPTQKATE
Mflv_1664|M.gilvum_PYR-GCK VIVTHNMQQAARVSDQTAFFNLEATGKPGRLIEIDDTEKIFSNPTQKATE
Mvan_5089|M.vanbaalenii_PYR-1 VIVTHNMQQAARVSDQTAFFNLEATGKPGRLIEVDDTEKIFSNPSQKATE
MMAR_4860|M.marinum_M VIVTHNMQQAARVSDQTAFFNLEAVGKPGRLIEIDDTEKIFSNPREKSTE
MUL_0440|M.ulcerans_Agy99 VIVTHNMQQAARVSDQTAFFNLEAVGKPGRLIEIDDTEKIFSNPREKSTE
MAV_0767|M.avium_104 VIVTHNMQQAARVSDYTAFFNLEAVGKPGRLIEVDDTEKIFSNPSQKATE
Mb0843|M.bovis_AF2122/97 VIVTHNMQQAARVSDQTAFFNLEAVGKPGRLVEIASTEKIFSNPNQKATE
Rv0820|M.tuberculosis_H37Rv VIVTHNMQQAARVSDQTAFFNLEAVGKPGRLVEIASTEKIFSNPNQKATE
MLBr_02189|M.leprae_Br4923 VIVTHNMQQAARVSDQTAFFNLEAVGRPGRLVEIDDTEKIFSNPTEKATE
*************** ********.*:** *:*: .******** :*:**
MSMEG_5779|M.smegmatis_MC2_155 DYISGRFG
TH_2660|M.thermoresistible__bu DYISGRFG
MAB_0749|M.abscessus_ATCC_1997 DYISGRFG
Mflv_1664|M.gilvum_PYR-GCK DYISGRFG
Mvan_5089|M.vanbaalenii_PYR-1 DYISGRFG
MMAR_4860|M.marinum_M DYISGRFG
MUL_0440|M.ulcerans_Agy99 DYISGRFG
MAV_0767|M.avium_104 DYISGRFG
Mb0843|M.bovis_AF2122/97 DYISGRFG
Rv0820|M.tuberculosis_H37Rv DYISGRFG
MLBr_02189|M.leprae_Br4923 DYISGRFG
********