For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADAHVSIARRRSGPPRGRRRALPILLRTGAAALGSTVAGLGYAAIIERNAFVLREITMPVLSPGSTPLRV LHISDLHMTPGQRRKQAWLRELAGWEPDLVVNTGDNLAHPKAVPAVIQALGDLLSRPGVFVFGSNDYFGP HLKNPLNYVTNPSHRVRGEPLPWQDLRAAFTERGWLDLTHTRREFEVAGLHVAAAGVDDPHIDRDRYDTI AGPASPAANLRLGLTHSPEPRVLDRFAADGYQLVMAGHTHGGQVCLPFYGALVTNCGLDRSRAKGPSRWG AHMQLHVSAGMGTSPYAPVRFCCRPEATLLTLIATPMGDRDAVSDLSRSQPTASVR*
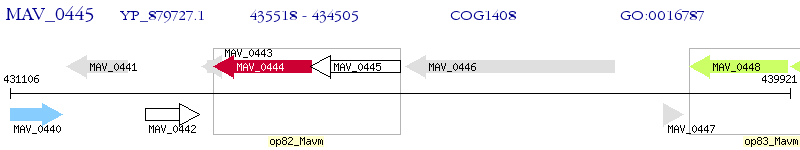
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0445 | - | - | 100% (337) | secreted protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3708 | - | 1e-167 | 87.30% (315) | hypothetical protein Mb3708 |
| M. gilvum PYR-GCK | Mflv_1364 | - | 1e-149 | 78.27% (313) | metallophosphoesterase |
| M. tuberculosis H37Rv | Rv3683 | - | 1e-167 | 87.30% (315) | hypothetical protein Rv3683 |
| M. leprae Br4923 | MLBr_02309 | - | 1e-157 | 86.49% (296) | hypothetical protein MLBr_02309 |
| M. abscessus ATCC 19977 | MAB_0407c | - | 1e-136 | 71.57% (313) | hypothetical protein MAB_0407c |
| M. marinum M | MMAR_5172 | - | 1e-171 | 84.27% (337) | hypothetical protein MMAR_5172 |
| M. smegmatis MC2 155 | MSMEG_6202 | - | 1e-149 | 78.23% (317) | secreted protein |
| M. thermoresistible (build 8) | TH_1706 | - | 1e-148 | 79.68% (310) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4258 | - | 1e-171 | 84.27% (337) | hypothetical protein MUL_4258 |
| M. vanbaalenii PYR-1 | Mvan_5443 | - | 1e-156 | 81.15% (313) | metallophosphoesterase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6202|M.smegmatis_MC2_155 ---------MSAVSLVKTAVKTAPVIKKAAAITAGSLVAGVGYASLIERN
TH_1706|M.thermoresistible__bu ---------MPVAS-------SRSVARKSAALAVGSLAAGIGYASLIERN
Mflv_1364|M.gilvum_PYR-GCK ---------MPSHS-------PGAILKTTALASAGTLAAGIAYASLIERN
Mvan_5443|M.vanbaalenii_PYR-1 ------MPSAPSGS-------PGSILKATAAASVGTLVAGIGYASLIERN
MAB_0407c|M.abscessus_ATCC_199 ---------MSTS--------LNTVIKRAAAYSAGSAALGIGYASIIERN
Mb3708|M.bovis_AF2122/97 -------MAAVLP-----------TLIRTGAVALGSAIAGIGYAALVERN
Rv3683|M.tuberculosis_H37Rv -------MAAVLP-----------TLIRTGAVALGSAIAGIGYAALVERN
MMAR_5172|M.marinum_M MAYAQPPITKLLPGPLSGRHRAASVLLRGSAVALGTTVAGIGYASLIERN
MUL_4258|M.ulcerans_Agy99 MAYAQPPITKLLPGPLSGRHRAASVLLRGSAVALGTTVAGIGYASLIERN
MAV_0445|M.avium_104 MADAHVSIARRRSGPPRGRRRALPILLRTGAAALGSTVAGLGYAAIIERN
MLBr_02309|M.leprae_Br4923 -------------------------MIRTGALVLSSAATGVGYASIVERN
. .: *:.**:::***
MSMEG_6202|M.smegmatis_MC2_155 AFVLREATMPVLAPGSTPLRVLHISDLHMRPNQRRKQAWLRDLARLEPDL
TH_1706|M.thermoresistible__bu AFTLREVTMPVLAPGSTPLKVLHISDIHMRPTQRRKQAWLRELARLEPDL
Mflv_1364|M.gilvum_PYR-GCK AFVVREATMPVLTPGSSPLKVLHISDIHMRPQQRRKQEWLRELAQWEPDF
Mvan_5443|M.vanbaalenii_PYR-1 AFVVRETTMPVLSPGSSPLKVLHISDLHMRPSQRRKQAWVRDLARWQPDF
MAB_0407c|M.abscessus_ATCC_199 AFTVREATMDVLDPGASPLRVLHISDIHMRPGQRLKQAWLRELAQWQPDL
Mb3708|M.bovis_AF2122/97 AFVLREVTMPVLTPGSTPLRVLHISDLHMLPNQHRKQAWLRELASWEPDL
Rv3683|M.tuberculosis_H37Rv AFVLREVTMPVLTPGSTPLRVLHISDLHMLPNQHRKQAWLRELASWEPDL
MMAR_5172|M.marinum_M AFVLREVTMPVLSPGSTPLRVLHLSDLHMLPNQRRKQAWLRELASWEPDL
MUL_4258|M.ulcerans_Agy99 AFVLREVTMPVLSPGSTPLRVLHLSDLHMLPNQRRKQAWLRELASWEPDL
MAV_0445|M.avium_104 AFVLREITMPVLSPGSTPLRVLHISDLHMTPGQRRKQAWLRELAGWEPDL
MLBr_02309|M.leprae_Br4923 AFVLRELTIPILTPGSTPLRVLHISDIHMMPHQRRKQAWLRDLATWQPDL
**.:** *: :* **::**:***:**:** * *: ** *:*:** :**:
MSMEG_6202|M.smegmatis_MC2_155 VVNTGDNLAHPKAVPAVVQSLSELLSVPGLFVFGSNDYFGPRPKNPLNYV
TH_1706|M.thermoresistible__bu VVNTGDNLAHPKAVPTVVQALNPLLSLPGLFVFGSNDYFAPRLKNPLNYV
Mflv_1364|M.gilvum_PYR-GCK VVNTGDNLSHPKAVPAVVQALGDLLAVPGVFVFGSNDYFAPKPKNPANYL
Mvan_5443|M.vanbaalenii_PYR-1 VVNTGDNLAHPKAVPSVVQALGDLLSVPGVFVFGSNDYFAPRLKNPANYL
MAB_0407c|M.abscessus_ATCC_199 VINTGDNLAHPKAVSSVVQSLGDLLSIPGLFVFGSNDYFGPRLKNPAKYL
Mb3708|M.bovis_AF2122/97 VVNTGDNLAHPKAVPAVVQTLSDLLSRPGVFVFGSNDYFGPRLKNPMNYL
Rv3683|M.tuberculosis_H37Rv VVNTGDNLAHPKAVPAVVQTLSDLLSRPGVFVFGSNDYFGPRLKNPMNYL
MMAR_5172|M.marinum_M VVNTGDNLAHPKAVPAVVQTLGDLLSRPGVFVFGSNDYFGPRLKNPANYL
MUL_4258|M.ulcerans_Agy99 VVNTGDNLAHPKAVPAVVQTLGDLLSRPGVFVFGSNDYFGPRLKNPANYL
MAV_0445|M.avium_104 VVNTGDNLAHPKAVPAVIQALGDLLSRPGVFVFGSNDYFGPHLKNPLNYV
MLBr_02309|M.leprae_Br4923 VVNTGDNLAHPKAVPGVIQALGNLLSRPGVFVFGSNDYFGPHLKNPVNYL
*:******:*****. *:*:*. **: **:*********.*: *** :*:
MSMEG_6202|M.smegmatis_MC2_155 TKPQHRTQGEALPWQDLRAAFTERGWLDLTHTRRDLEVAGLHLSVAGVDD
TH_1706|M.thermoresistible__bu VKPSHRVFGEPLPWQDLRAAFTERGWLDLTHTRRVIEVAGLRIAAAGVDD
Mflv_1364|M.gilvum_PYR-GCK TKPGRRIHGEPLPWQDLRAAFTERGWLDMTHTRREFEVKGLHIAAAGVDD
Mvan_5443|M.vanbaalenii_PYR-1 TNPGHRIHGEPLPWQDLRAAFTERGWLDMTHTRREFEVAGLHIAAAGVDD
MAB_0407c|M.abscessus_ATCC_199 YKSHERKHGEPLPWQDLRAAFTERGWHDMTHTRREIEVGGVRIAAAGVDD
Mb3708|M.bovis_AF2122/97 TSPDHRVRGAALPWQDLRAAFTERGWLDLTHTRREFEVAGLHIAAAGVDD
Rv3683|M.tuberculosis_H37Rv TSPDHRVRGAALPWQDLRAAFTERGWLDLTHTRREFEVAGLHIAAAGVDD
MMAR_5172|M.marinum_M INPGHRVRGEPLPWQDLRAAFTERGWLDLTHTRREFEVAGLHIAAAGVDD
MUL_4258|M.ulcerans_Agy99 INPGHRVRGEPLPWQDLRAAFTERGWLDLTHTRREFEVAGLHIAAAGVDD
MAV_0445|M.avium_104 TNPSHRVRGEPLPWQDLRAAFTERGWLDLTHTRREFEVAGLHVAAAGVDD
MLBr_02309|M.leprae_Br4923 TNPTHRVRGKPLPWKDLRAAFTERGWLDLTHTRREFEVAGLHIAAAGVDD
.. .* * .***:*********** *:***** :** *::::.*****
MSMEG_6202|M.smegmatis_MC2_155 PHLKRDRYDTVAGPANAAANLTLGLTHSPEPRVLDRFAADGYQLVLAGHT
TH_1706|M.thermoresistible__bu PHLKRDRYPTIAGPADRTANLTLGLTHSPEPRVLDRFAADGYQLVLAGHT
Mflv_1364|M.gilvum_PYR-GCK PHLKRDRYDTVAGPASPAANLTLGLTHSPEPRVLDRFAADGYQLVMAGHT
Mvan_5443|M.vanbaalenii_PYR-1 PHLKRDRYDTIAGPASPTANLTLGLTHSPEPRVLDRFAADGYQLVMAGHT
MAB_0407c|M.abscessus_ATCC_199 PHLHRDRYDTIAGMPNPLATLRLGLTHSPEPRVLDRFAEDGYQLVMAGHT
Mb3708|M.bovis_AF2122/97 PHIDRDRYDTIAGPASPAANLRLGLTHSPEPRVLDRFAADGYQLVLAGHT
Rv3683|M.tuberculosis_H37Rv PHIDRDRYDTIAGPASPAANLRLGLTHSPEPRVLDRFAADGYQLVLAGHT
MMAR_5172|M.marinum_M PHINRDRYDAIAGPASPAANLRLGLTHSPEPRVLDRFAADGYQLVMAGHT
MUL_4258|M.ulcerans_Agy99 PHINRDRYDAIAGPASPAANLRLGLTHSPEPRVLDRFAADGYQLVMAGHT
MAV_0445|M.avium_104 PHIDRDRYDTIAGPASPAANLRLGLTHSPEPRVLDRFAADGYQLVMAGHT
MLBr_02309|M.leprae_Br4923 PHISRDRYDTIAGPASPVANLRLGLTHSPEPRVLDRFAADCYQLVMAGHT
**: **** ::** .. *.* **************** * ****:****
MSMEG_6202|M.smegmatis_MC2_155 HGGQLCLPFYGALVTNCELDRSRAKGASRWGADMQLHVSAGIGTSPYAPV
TH_1706|M.thermoresistible__bu HGGQLCLPFYGALVTNCELDRSRAKGASRWGANMALHVSAGIGTSPYAPV
Mflv_1364|M.gilvum_PYR-GCK HGGQLCLPFYGAIVTNCDLDRSRAKGPSRWGVRTQLHVSAGIGTSPYAPL
Mvan_5443|M.vanbaalenii_PYR-1 HGGQLCLPFYGAIVTNCDLDRSRAKGPSRWGARTQLHVSAGLGTSPYAPL
MAB_0407c|M.abscessus_ATCC_199 HGGQLCLPLYGAIVTNCELDRSRVKGPSQWGSHMQLHVSAGIGTSPFAPM
Mb3708|M.bovis_AF2122/97 HGGQLCLPLYGALVTNCGLDRSRAKGASHWGANMRLHVSAGIGTSPFAPV
Rv3683|M.tuberculosis_H37Rv HGGQLCLPLYGALVTNCGLDRSRAKGASHWGANMRLHVSAGIGTSPFAPV
MMAR_5172|M.marinum_M HGGQLCLPFYGALVTNCGLDRTRAKGASQWGANMRLHVSAGIGTSPFAPV
MUL_4258|M.ulcerans_Agy99 HGGQLCLPFYGALVTNCGLDRTRAKGASQWGANMRLHVSAGIGTSPFAPV
MAV_0445|M.avium_104 HGGQVCLPFYGALVTNCGLDRSRAKGPSRWGAHMQLHVSAGMGTSPYAPV
MLBr_02309|M.leprae_Br4923 HGGQLCLPFYGALVTNCGLDRSRAKGLSRWDADMRLHVSAGIGTSPLAPM
****:***:***:**** ***:*.** *:*. ******:**** **:
MSMEG_6202|M.smegmatis_MC2_155 RFCCRPEATLLTLVAAPTGGGNAG---TRAGTSSPSVSAR----------
TH_1706|M.thermoresistible__bu RFCCRPEATLLTLVAAPSGGADSG---SHAGRAHPTVSAQKTTG------
Mflv_1364|M.gilvum_PYR-GCK RFCCRPEATMLTLVAAPTGGSDMG---SRAGLSHPTVSAR----------
Mvan_5443|M.vanbaalenii_PYR-1 RFCCRPEATMLTLVAAPTGGSDVG---SRAGHSSPTVSAR----------
MAB_0407c|M.abscessus_ATCC_199 RFFCRPEATLLTLVPARTSGHGAD---AESGDRSPSATVQ----------
Mb3708|M.bovis_AF2122/97 RFCCRPEATLLTLIATPMGGRDSS---SNLGRSQPTVSVR----------
Rv3683|M.tuberculosis_H37Rv RFCCRPEATLLTLIATPMGGRDSS---SNLGRSQPTVSVR----------
MMAR_5172|M.marinum_M RFCCRPEATLLTLIAAPMGGRDSS---SNLDRSQPTMSIR----------
MUL_4258|M.ulcerans_Agy99 RFCCRPEATLLTLIAAPMGGRDSS---SNLDRSQPTMSIR----------
MAV_0445|M.avium_104 RFCCRPEATLLTLIATPMGDRDAV---SDLSRSQPTASVR----------
MLBr_02309|M.leprae_Br4923 RFCCRPEATLLTLIATPMGSRVESPVRTGAVRNQQLQCVERAHRDSGPRP
** ******:***:.: .. : .
MSMEG_6202|M.smegmatis_MC2_155 -----
TH_1706|M.thermoresistible__bu -----
Mflv_1364|M.gilvum_PYR-GCK -----
Mvan_5443|M.vanbaalenii_PYR-1 -----
MAB_0407c|M.abscessus_ATCC_199 -----
Mb3708|M.bovis_AF2122/97 -----
Rv3683|M.tuberculosis_H37Rv -----
MMAR_5172|M.marinum_M -----
MUL_4258|M.ulcerans_Agy99 -----
MAV_0445|M.avium_104 -----
MLBr_02309|M.leprae_Br4923 PTQLD