For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RLVADSGLWTTGRADVTEEPPLVALLEVSGAVLSWTLDDPDDAPPRLTFTDVSRADWLWRVLGEAGHVAL VSAVRDAAVPAGGIELAGVDLAPGALAPLRRLATGHWLRRWWPASRQDGIAGLDRALLDAELALLTAGAQ HFFPDDTLDSDVAGLLAPHATALLGHLVSPDPRVADLVRASAGLADGIGVDAGRWPELLAALDDSSLVVA PPSDRRDDYALAAGAGAGPRAAAIARGVASVAWSGVPPGVFDAAEDTVGWIIRVDGSAVVAVVQADVIGP DPATGIAVRLRSAAVEGAGALDAAGRATFPLTGARGALTESAAWGHDWSATELQVGAAVSTESRHTRERV RRWARARLESPPRDAFLAERLAAESAY*
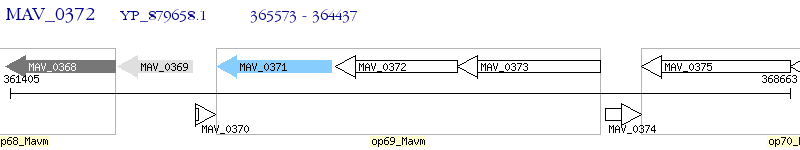
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0372 | - | - | 100% (378) | hypothetical protein MAV_0372 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4069c | - | 9e-99 | 53.16% (380) | hypothetical protein MAB_4069c |
| M. marinum M | MMAR_5249 | - | 1e-133 | 64.34% (387) | hypothetical protein MMAR_5249 |
| M. smegmatis MC2 155 | MSMEG_3798 | - | 3e-88 | 52.51% (358) | hypothetical protein MSMEG_3798 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4325 | - | 1e-132 | 63.57% (387) | hypothetical protein MUL_4325 |
| M. vanbaalenii PYR-1 | Mvan_3333 | - | 2e-99 | 55.15% (379) | hypothetical protein Mvan_3333 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5249|M.marinum_M MKLVAESGLWSTGATGPDEQLPASPLIALLEVSGAVLSWVIDDPRGEAAT
MUL_4325|M.ulcerans_Agy99 MKLVAESGLWSTGATGPDEQLPASPLIALLEVSGAVLSWVIDDPRGEAAT
MAV_0372|M.avium_104 MRLVADSGLWTTGRADVTE---EPPLVALLEVSGAVLSWTLDDP-DDAPP
Mvan_3333|M.vanbaalenii_PYR-1 MRFAEDSGLWTTG-PGPDA----VQLSAVLEVSGAVLSWSVDGP--ESDV
MAB_4069c|M.abscessus_ATCC_199 MRLVNEAGLWTTG-PAPAP----VPLVAVLEVSGAVLSWPVDD--PSQPP
MSMEG_3798|M.smegmatis_MC2_155 ----------MTD-PGYPP----LPSVAVLEVSGAVLSWIVDEE-PSTAP
*. . *:********** :* .
MMAR_5249|M.marinum_M RITFTNAARADWLWRVVGESGHVAVLSAV--AGHPAGDVDLRGVDLIPGS
MUL_4325|M.ulcerans_Agy99 RITFTDAARADWLWRVVGESGHVAVLSAV--AGHPAGDVDLRGVDLIPGS
MAV_0372|M.avium_104 RLTFTDVSRADWLWRVLGEAGHVALVSAVRDAAVPAGGIELAGVDLAPGA
Mvan_3333|M.vanbaalenii_PYR-1 HIAFTDPVRADWLWRVVGESAHSAIVSAL--GGVPDGQVEVEGVAVRPAA
MAB_4069c|M.abscessus_ATCC_199 VVSFTDAMRADWMWRVFGEAGHVAVVEALR-DAAPGKAIELPSVSVLPRP
MSMEG_3798|M.smegmatis_MC2_155 EISFTDPLRADWLWRVVGEQAHVAVVEGR-----------TADLQVLPGS
::**: ****:***.** .* *::.. .: : * .
MMAR_5249|M.marinum_M AAGLRRLAVGHWLRRWWPASQRDDIAGLDRALLDVEVALLTVGAQGFFTD
MUL_4325|M.ulcerans_Agy99 AAGLRRLAVGHWLRRWWPASQRDDIAGLDRALLDVEVALLTVGAQGFFTD
MAV_0372|M.avium_104 LAPLRRLATGHWLRRWWPASRQDGIAGLDRALLDAELALLTAGAQHFFPD
Mvan_3333|M.vanbaalenii_PYR-1 LDSLRRLALGHWMRRWWPASVRDGIPELGAAVLDGELALLTAAAGDYFVD
MAB_4069c|M.abscessus_ATCC_199 ADSLRRLALGHWLRRWWPASLREGVGPLDRAVLDAEVAVLTATAEDYFTD
MSMEG_3798|M.smegmatis_MC2_155 TDALRRLALGHWLRRWWPASIREGIAELDPALLDAEIALSTVAAEEFFTD
***** ***:******* ::.: *. *:** *:*: *. * :* *
MMAR_5249|M.marinum_M DTLDSDVADLLAPHAGALTAHVRTDDARIRALVRAGAHLADEVGADGAGW
MUL_4325|M.ulcerans_Agy99 DTLDSDVAELLAPHAGALTAHVRTDDARIRALVRAGAHLAAEVGTDGAGW
MAV_0372|M.avium_104 DTLDSDVAGLLAPHATALLGHLVSPDPRVADLVRASAGLADGIGVDAGRW
Mvan_3333|M.vanbaalenii_PYR-1 DTFDSDVAELLRPHAELLEGLARLGDPRVAELARACAELAADIGVEAG--
MAB_4069c|M.abscessus_ATCC_199 DTLDADVAGLLAPHAA---ALDSHRDPRVGLLAARCRDLADEMG------
MSMEG_3798|M.smegmatis_MC2_155 DTLDSDVAGLLAPHTARLDALAAEGDPRVTALIDRCRDLAAEIG------
**:*:*** ** **: . *.*: * ** :*
MMAR_5249|M.marinum_M TELSAAIDDSSVAVTMPTGRRDDYALAAGAGQGPRGAASIGRGVASINWG
MUL_4325|M.ulcerans_Agy99 TELSAAIDDSSVAVTMPTGRRDDYALAAGAGQGPRGAASIGRGVASINWG
MAV_0372|M.avium_104 PELLAALDDSSLVVAPPSDRRDDYALAAGAGAGPR-AAAIARGVASVAWS
Mvan_3333|M.vanbaalenii_PYR-1 --------PAAVAVQ---SRRDDYALVAG-EVTRRGAAAIAAGVASVSWA
MAB_4069c|M.abscessus_ATCC_199 -------LQWDVR-DAEVARREDYALAAGAGDERGAPGTIAMGTATIAWS
MSMEG_3798|M.smegmatis_MC2_155 -------VDWTATVPVGPVRRADYALAAGSRTSR--QDAIAGGVATVRWS
** ****.** :*. *.*:: *.
MMAR_5249|M.marinum_M GVPPAIFDAAEDTVDWTIEPGGPAGSAVVAVVRAALIGAQPATDVAVRVR
MUL_4325|M.ulcerans_Agy99 GVPPAIFDAAEDTVDWTIEPGGPAGSAVVAVVRAALIGAQPATDVPVRVR
MAV_0372|M.avium_104 GVPPGVFDAAEDTVGWIIRVDG---SAVVAVVQADVIGPDPATGIAVRLR
Mvan_3333|M.vanbaalenii_PYR-1 AVPPGVFDAAENTVSWRVESAD---DVANAVVLVELSGRASAAGIAVRVR
MAB_4069c|M.abscessus_ATCC_199 DVPPGIFDAAEHNLDWAVVSDG---ATVTIRLRAAICGPGSALDLPITLH
MSMEG_3798|M.smegmatis_MC2_155 AVPPGIFDAAEDTVEWSVRSDG---GAVDVVVQVALSGPGHADGIPVTVR
***.:*****..: * : . .. : . : * * .:.: ::
MMAR_5249|M.marinum_M SGEVSGTGALDAGGRATVALVDAQRRAMTQTSAWDHNWAATSVVIGA-DI
MUL_4325|M.ulcerans_Agy99 SGEASGTGALDAGGRATVALVDAQRRAMTQTSAWDHNWAATSVVIGA-DI
MAV_0372|M.avium_104 SAAVEGAGALDAAGRATFPLTGARG-ALTESAAWGHDWSATELQVGA-AV
Mvan_3333|M.vanbaalenii_PYR-1 SGAVDGAGTLDASGAATVALVGEDQAPITESVAWGRDWRDTAVTIGA-DV
MAB_4069c|M.abscessus_ATCC_199 CGVFQAIGVLDAEGAATLPVLDTNGQPAGEMQAWNADWSVAEVSVGAGRA
MSMEG_3798|M.smegmatis_MC2_155 CGDHGGAGVLAADGRAEIPMV-TGGRPTTETEAWNQDWSTAVVHVGATAG
.. . *.* * * * ..: . : **. :* : : :**
MMAR_5249|M.marinum_M A-ESRQTRDRVRRWVRDRLDRPPPDAFLAEVLAAESAY
MUL_4325|M.ulcerans_Agy99 A-ESRQTRDRVRRWVRDRLDRPPPDAFLAEILASESAY
MAV_0372|M.avium_104 STESRHTRERVRRWARARLESPPRDAFLAERLAAESAY
Mvan_3333|M.vanbaalenii_PYR-1 G-ESPQVRQRVRTFARARLARPGPDAFLAELLAAESDY
MAB_4069c|M.abscessus_ATCC_199 AD-SSESRDRVRRFARARLAVPPADAFLAELLAAESDY
MSMEG_3798|M.smegmatis_MC2_155 PDEDARTRARVRAFARARLAAPTADAYLAEILAAESDY
. . * *** :.* ** * **:*** **:** *