For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTITLYTQPGCGGCIFAAKDLTKAGVPFTERNVRADPDAADMVHRLYQEHRDPGVVPETPVIVIDDEVFF GAVELHAYLRELGRESAA
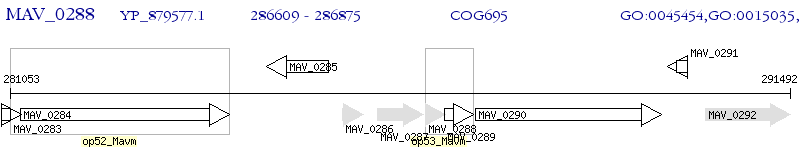
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0288 | - | - | 100% (88) | glutaredoxin |
| M. avium 104 | MAV_3921 | - | 6e-05 | 39.13% (46) | glutaredoxin |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4661 | - | 6e-05 | 41.46% (41) | glutaredoxin-like protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01736 | nrdH | 0.0001 | 36.96% (46) | glutaredoxin electron transport component of NrdEF |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1640 | nrdH | 2e-05 | 33.80% (71) | glutaredoxin electron transport component of NRDEF |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1876 | nrdH | 2e-05 | 33.80% (71) | glutaredoxin electron transport component of NRDEF |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1640|M.marinum_M -----MTITVYTKPACVQCSATYKALDKQGIDYDKVDISLDPEARDYVMA
MUL_1876|M.ulcerans_Agy99 -----MTITVYTKPACVQCSATYKALDKQGINYDKVDISLDPEARDYVMA
MLBr_01736|M.leprae_Br4923 ----MMNITVYTKPACVQCSATYKALDKQGIPYQTVDISLDSEARDYVMA
MAV_0288|M.avium_104 -----MTITLYTQPGCGGCIFAAKDLTKAGVPFTERNVRADPDAADMVHR
Mflv_4661|M.gilvum_PYR-GCK MSTDVPAVTMYTTSWCGYCVRLKKVLKNEGITFAEVNIEEDPAAAEFVGS
:*:** . * * * * : *: : :: *. * : *
MMAR_1640|M.marinum_M L-------GYL-QAPVVVAGDDHWSGFRP--DRIKALAKSALSA
MUL_1876|M.ulcerans_Agy99 L-------GYL-QAPVVVAGDDHWSGFRP--DRIKALAKSALSA
MLBr_01736|M.leprae_Br4923 L-------GYL-QAPVVVAGNNHWSGFRP--DRIKALAGAVLTA
MAV_0288|M.avium_104 LYQEHRDPGVVPETPVIVIDDEVFFGAVELHAYLRELGRESAA-
Mflv_4661|M.gilvum_PYR-GCK VNG-----GNHVVPTLKFADGSTLTNPTG--AQVKAKLAS----
: * ..: . ... . ::