For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RSALATLGQMAVAVLVAVVVAVVSLTAIAGVQWPAFPSSNQLHALTTVGQVGCLAGLVAVGWVWHSFTGR VRRLAQLGGLVFVSAFTVVTLGMPLGATKLYLFGISVDQQFRTEYLTRLADSPALRDMTYLGLPPFYPPG WFWIGGRAAALSGTPAWEVFKPWAITSITVAVAVALVLWWRMVRFEYALIVTTATAAVTLAYGSPEPYSA MITVLLPPMLVLTWSGLRAGECRAGGMPETAERAAGGAPETAERETNFTVGAERETNFTLAAGEAPSAPR ETPSAPGETPRSGWAAVVGAGLFLGFTATWYTLLFGFTAFAVGLMALWLAGLRWRRHGVRAALEPLRRLA VIVGIAVAIGCTTWLPFLVRAARNPVSNTGSAQHYLPADGAELTFPMLQFTLLGVVCMAGTLWLVLRARA SVRAGALAIGVLAVYLWSLLSMLTTLARTTLLSFRLQPTLSVLLVAAGAFGLVEAALALRRRSRAVIPVA GAIGLAAAIAFSQDIPDVLRPDLTIAYTDTDGHGQRGDRRPPSAEKFYASIDDVITRETGRPRDQTVVMT ADYSFLSFYPYWGFQGLTSHYANPLAQFDLRAAQIDKWSKLKTADELLHALDTCPWPPPTVFLMRHGANN TYTLRLAEDVYPNQPNVRRYTVELRAALFDSPRFAVHRVGPFVLAVRKPAA*
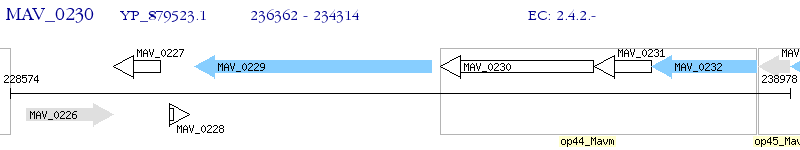
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0230 | - | - | 100% (682) | hypothetical protein MAV_0230 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3821 | - | 0.0 | 66.86% (679) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_1176 | - | 0.0 | 55.91% (685) | putative transmembrane protein |
| M. tuberculosis H37Rv | Rv3792 | - | 0.0 | 66.86% (679) | transmembrane protein |
| M. leprae Br4923 | MLBr_00107 | - | 0.0 | 72.21% (680) | hypothetical protein MLBr_00107 |
| M. abscessus ATCC 19977 | MAB_0190c | - | 0.0 | 57.94% (680) | hypothetical protein MAB_0190c |
| M. marinum M | MMAR_5354 | - | 0.0 | 74.78% (686) | hypothetical protein MMAR_5354 |
| M. smegmatis MC2 155 | MSMEG_6386 | - | 1e-141 | 64.39% (396) | membrane protein |
| M. thermoresistible (build 8) | TH_1348 | - | 0.0 | 61.42% (591) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4971 | - | 0.0 | 73.91% (686) | hypothetical protein MUL_4971 |
| M. vanbaalenii PYR-1 | Mvan_5629 | - | 0.0 | 60.47% (678) | putative transmembrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1176|M.gilvum_PYR-GCK ---------------MRG------ALATLGQMLVGAVVAAVVATVSLIAI
Mvan_5629|M.vanbaalenii_PYR-1 ---------------MRAGDTLAAPVRVAGQMLVATVVAVAVATVSLVAI
MSMEG_6386|M.smegmatis_MC2_155 -----------------------MAARVLGQMVLAVLMASAVAGVAIAAI
TH_1348|M.thermoresistible__bu --------------------------------------------------
MAV_0230|M.avium_104 ---------------------MRSALATLGQMAVAVLVAVVVAVVSLTAI
MLBr_00107|M.leprae_Br4923 ---------------------MRNALASFGQIVLAAVVASGVAAVSLIAI
Mb3821|M.bovis_AF2122/97 MPSRRKSPQFGHEMGAFTSARAREVLVALGQLAAAVVVAVGVAVVSLLAI
Rv3792|M.tuberculosis_H37Rv MPSRRKSPQFGHEMGAFTSARAREVLVALGQLAAAVVVAVGVAVVSLLAI
MMAR_5354|M.marinum_M ---------------------MRNALATLGQMAAAVLVAVTVAVVALSAI
MUL_4971|M.ulcerans_Agy99 ---------------------MRNALATLGQMAAAVLVAVTVAVVALSAI
MAB_0190c|M.abscessus_ATCC_199 ------------MTAQTAEAARRHVLDTTKDLTLAVVTAAAVSVVALTAI
Mflv_1176|M.gilvum_PYR-GCK ARVEWPAYNSSNQLHALTTVGQVACLLGLLAAGILWRR---GRQTVARIG
Mvan_5629|M.vanbaalenii_PYR-1 ARVEWPAYNSSNQLHALTTVGQVACLIGLLASGLAWRR---GRTVLARLG
MSMEG_6386|M.smegmatis_MC2_155 ARVEWPAYNTSNQLHALTTVGQFGCLAGLFAAGLLWRR---GRRTLARLG
TH_1348|M.thermoresistible__bu --------------------------------------------------
MAV_0230|M.avium_104 AGVQWPAFPSSNQLHALTTVGQVGCLAGLVAVGWVWHSFTGRVRRLAQLG
MLBr_00107|M.leprae_Br4923 ARVHWPAFPSSNQLHALTTVGQVGCLTGLLAVGGVWQ--AGRFRRLAQLG
Mb3821|M.bovis_AF2122/97 ARVEWPAFPSSNQLHALTTVGQVGCLAGLVGIGWLWR--HGRFRRLARLG
Rv3792|M.tuberculosis_H37Rv ARVEWPAFPSSNQLHALTTVGQVGCLAGLVGIGWLWR--HGRFRRLARLG
MMAR_5354|M.marinum_M SQVQWPAFPSSNQLHALTTVGQVGCLAGLLGAGWAWRR-GGRLRRPAQLG
MUL_4971|M.ulcerans_Agy99 SQVQWPAFPSSNQLHALTTVGQVGCLAGLLGAGWAWRR-GGRLRRPAQLG
MAB_0190c|M.abscessus_ATCC_199 ARVEWPAFGSSNQLHALTTVGQFGAIAGLVLAGVLWRR---GHRAVSKWL
Mflv_1176|M.gilvum_PYR-GCK SALFLSAFTVVTLAMPLGATKLYLFGISVDQQFRTEYLTRLADEPGLRDM
Mvan_5629|M.vanbaalenii_PYR-1 APVFLSAFAVVTLAMPLGATKLYLFGISVDQQFRTEYMTRFADQPGLHDM
MSMEG_6386|M.smegmatis_MC2_155 ALAFISAFSVVTLAMPLGATKLYLFGVSVDQQFRTEYLTRLTDTAGLHDM
TH_1348|M.thermoresistible__bu --------------MPLGATKLYLFGISVDQQFRTEYLTRFADTAALRDM
MAV_0230|M.avium_104 GLVFVSAFTVVTLGMPLGATKLYLFGISVDQQFRTEYLTRLADSPALRDM
MLBr_00107|M.leprae_Br4923 GLVFVSAFTVVTLGMPLGATKLYLFGISVDQQFRTEYLTRLTDSAALQDM
Mb3821|M.bovis_AF2122/97 GLVLVSAFTVVTLGMPLGATKLYLFGISVDQQFRTEYLTRLTDTAALRDM
Rv3792|M.tuberculosis_H37Rv GLVLVSAFTVVTLGMPLGATKLYLFGISVDQQFRTEYLTRLTDTAALRDM
MMAR_5354|M.marinum_M GLVFVSAFTVVTLGMPLGATRLYLFGISVDQQFRTEYLTRLTDSPALHDM
MUL_4971|M.ulcerans_Agy99 GLVFVSAFTVVTLGMPLGATRLYLFGISVDQQFRTEYLTRLTDSPALHDM
MAB_0190c|M.abscessus_ATCC_199 SLLFLATFTVVTVGMPLGATKLYLFGISVDQQFRTEYLTRFTDSPALHDM
******:*****:**********:**::* ..*:**
Mflv_1176|M.gilvum_PYR-GCK TYIGLPPFYPAGWFWLGGRLAALTGTPAWEMFKPWSIISITIAIVLAFVL
Mvan_5629|M.vanbaalenii_PYR-1 TYIGLPPFYPPGWFWIGGRLAALTGTPAWEMFKPWSVVSIAVAIVLAYAL
MSMEG_6386|M.smegmatis_MC2_155 TYIGLPPFYPAGWFWLGGRIAAATGTPAWEMFKPWSIVSITIAVALALVL
TH_1348|M.thermoresistible__bu TYLGLPPFYPPGWFWLGGRAAALTGIPAWEMFKPWSILSITIAAVLALVL
MAV_0230|M.avium_104 TYLGLPPFYPPGWFWIGGRAAALSGTPAWEVFKPWAITSITVAVAVALVL
MLBr_00107|M.leprae_Br4923 TYLGLPPFYPPGWFWIGGRVAALTGTPAWEIFKPWAITSITIAVAITLVL
Mb3821|M.bovis_AF2122/97 TYIGLPPFYPPGWFWIGGRAAALTGTPAWEMFKPWAITSMAIAVAVALVL
Rv3792|M.tuberculosis_H37Rv TYIGLPPFYPPGWFWIGGRAAALTGTPAWEMFKPWAITSMAIAVAVALVL
MMAR_5354|M.marinum_M TYIGLPPFYPPGWFWIGGRAAALTGSPAWEMFKPWAITSIAVAVAVALVL
MUL_4971|M.ulcerans_Agy99 IYIGLPPFYPPGWFWIGGRAAALTGSPAWEMFKPWAITSIAVAVAVALVL
MAB_0190c|M.abscessus_ATCC_199 TYPDMPPFYPAGWFWIGGRLAALTGTSGWEMYKPWAITSLAIAVCAAFIL
* .:*****.****:*** ** :* ..**::***:: *:::* : *
Mflv_1176|M.gilvum_PYR-GCK WAAMIRFEYALVVSTATAAAALAYSPAEPYAAIIAVLLPPVFVLAWSGLA
Mvan_5629|M.vanbaalenii_PYR-1 WAAMIRFEYALVVSTATAAATLAYSPAEPYAAIIAVLLPPVFVLAWSGLK
MSMEG_6386|M.smegmatis_MC2_155 WAAMIRFEYALVATAASTAAMLAYASTEPYAAIITVLMPPVFVLAWAGLR
TH_1348|M.thermoresistible__bu WAALVRFEYALIVTTATTAVMLAYSSTEPYAAIITVLLPPVFVLAWSGLR
MAV_0230|M.avium_104 WWRMVRFEYALIVTTATAAVTLAYGSPEPYSAMITVLLPPMLVLTWSGLR
MLBr_00107|M.leprae_Br4923 WWQMIRFEYALLVTIATAAVTLVYSSPEPYAAMITVLLPPALVLTWSGLR
Mb3821|M.bovis_AF2122/97 WWRMIRFEYALLVTVATAAVMLAYSSPEPYAAMITVLLPPMLVLTWSGLG
Rv3792|M.tuberculosis_H37Rv WWRMIRFEYALLVTVATAAVMLAYSSPEPYAAMITVLLPPMLVLTWSGLG
MMAR_5354|M.marinum_M WWRMIRFEYALLVTIATTAVTLAYSSPEPYAAMITVLLPPVLVLTWSGLR
MUL_4971|M.ulcerans_Agy99 WWRMIRFEYALLVTIATTAVTLAYSSPEPYAAMITVLLPPVLVLTWSGLR
MAB_0190c|M.abscessus_ATCC_199 WNRMIRFEYALIVSTATAAVTLAYASTEPYGAIVTVLLPPVLVLGYSALR
* ::******:.: *::*. *.*...***.*:::**:** :** ::.*
Mflv_1176|M.gilvum_PYR-GCK G----------------------RER------------------------
Mvan_5629|M.vanbaalenii_PYR-1 G----------------------RTRPGEPGIRR----------------
MSMEG_6386|M.smegmatis_MC2_155 A----------------------RTR------------------------
TH_1348|M.thermoresistible__bu G----------------------RTRNGGPPQAG----------------
MAV_0230|M.avium_104 AGECRAGGMPETAERAAGGAPETAERETNFTVGAERETNFTLAAGEAPSA
MLBr_00107|M.leprae_Br4923 A----------------------AEREADRTLGNKR--------------
Mb3821|M.bovis_AF2122/97 AR----------------------DRQ-----------------------
Rv3792|M.tuberculosis_H37Rv AR----------------------DRQ-----------------------
MMAR_5354|M.marinum_M AG-----------------APPAAERHASVAPDTESHADVTLG-------
MUL_4971|M.ulcerans_Agy99 AG-----------------APPAPERHASVAPDAESHADVTLG-------
MAB_0190c|M.abscessus_ATCC_199 ADR-----------------------------------------------
.
Mflv_1176|M.gilvum_PYR-GCK ------------SGGWAAVVGTGIFLGVAGLFYTLLFAYAAFTIAIMAVL
Mvan_5629|M.vanbaalenii_PYR-1 ------------SGGWAAIVGTGIFLGVTGLFYTLLLAYAAFTLTVMAVL
MSMEG_6386|M.smegmatis_MC2_155 ------------GGGWAAIVGVGIFLGVAALFYTLLLVYAAFTLTIMALC
TH_1348|M.thermoresistible__bu ------------GAPRGAVVGVGIFLGIAALFYTLLLAYAAFTLVIMAAV
MAV_0230|M.avium_104 PRETPSAPGETPRSGWAAVVGAGLFLGFTATWYTLLFGFTAFAVGLMALW
MLBr_00107|M.leprae_Br4923 --------------GWATVVGAGIFLGFAATWYTLLLAYTAFTVVLMTLL
Mb3821|M.bovis_AF2122/97 --------------GWAAVVGAGVFLGFAATWYTLLVAYGAFTVVLMALL
Rv3792|M.tuberculosis_H37Rv --------------GWAAVVGAGVFLGFAATWYTLLVAYGAFTVVLMALL
MMAR_5354|M.marinum_M ---------AAAARGWAAVVGAGIFLGFAATWYTLLVAYSAFTVTLMAAV
MUL_4971|M.ulcerans_Agy99 ---------AEAARGWGAVVGAGIFLGFAATWYTLLVAYSAFTVTLMAAL
MAB_0190c|M.abscessus_ATCC_199 ------------GAGWGAVVGTGVFLGFAALFYTLLTAFGALAITVAAAV
.::**.*:***.:. :**** : *::: : :
Mflv_1176|M.gilvum_PYR-GCK LAAARRH--------WDPLLRVVVIAVLSGLIMLLTWGPYLLAALRGTPA
Mvan_5629|M.vanbaalenii_PYR-1 LAAARRH--------WDPLLRLLTIAVISGLIALIGWGPYLLAAARGTPA
MSMEG_6386|M.smegmatis_MC2_155 VAVARRH--------IDPLLRLAVIAVISGAIALLTWAPYLLAAMRGEPA
TH_1348|M.thermoresistible__bu AAVTDRD--------WGPVRRLAVIAAVSIPIALIGWGPWLRAALHGEPA
MAV_0230|M.avium_104 LAGLRWRRHGVRA-ALEPLRRLAVIVGIAVAIGCTTWLPFLVRAARNPVS
MLBr_00107|M.leprae_Br4923 LATALCRRAGFRA-TFDPLRRLAGIVVIAAAIGAITWLPFLARAAHDPVS
Mb3821|M.bovis_AF2122/97 LAGSRLQ-SGIKA-AVDPLCRLAVVGAIAAAIGSTTWLPYLLRAARDPVS
Rv3792|M.tuberculosis_H37Rv LAGSRLQ-SGIKA-AVDPLCRLAVVGAIAAAIGSTTWLPYLLRAARDPVS
MMAR_5354|M.marinum_M LAVSRWRRGGLRA-AVDPLRRLAVIAAIAATIASTTWLPYLLRAARDPVS
MUL_4971|M.ulcerans_Agy99 LVVSRWRRGGLRA-AVDPLRRLAVIAVIAATIASTTWLPYLLRAARDPVS
MAB_0190c|M.abscessus_ATCC_199 VAMGRVRQNGWWPGVKEPLLRWLAIAAISIVIALVFWGPYLVAVAHASLE
. *: * : :: * * *:* . :
Mflv_1176|M.gilvum_PYR-GCK ESGTAQHYLPKDGAELSFPMLQFTLIGALCMLGTVWLVVRARSSTRAGAL
Mvan_5629|M.vanbaalenii_PYR-1 ETGTAQHYLPRDGAELSFPMLQFTLLGALCLLGTAWLVIYARSSTRAGAL
MSMEG_6386|M.smegmatis_MC2_155 DSGTAQHYLPDAGAELHFPMFSLTLHGALCMLGTVWLVVRARTSTRAGAL
TH_1348|M.thermoresistible__bu DTGTAQHYLPADGAELTFPMLDFTLVGALCMIGTIWLVARARNSARAAAL
MAV_0230|M.avium_104 NTGSAQHYLPADGAELTFPMLQFTLLGVVCMAGTLWLVLRARASVRAGAL
MLBr_00107|M.leprae_Br4923 DTGSAQHYLPADGAELAFPMLQFSLLGMICMLGTLWLIVRTSSSVRASAL
Mb3821|M.bovis_AF2122/97 DTGSAQHYLPADGAALTFPMLQFSLLGAICLLGTLWLVMRARSSAPAGAL
Rv3792|M.tuberculosis_H37Rv DTGSAQHYLPADGAALTFPMLQFSLLGAICLLGTLWLVMRARSSAPAGAL
MMAR_5354|M.marinum_M DTGSAQHYLPADGAELTFPMLQFSLLGALCMLGTLWLVVRARSSTRAGAL
MUL_4971|M.ulcerans_Agy99 DTGSAQHYLPADGAELTFPMLQFSLLGALCMLGTLWLVVRARSSTRAGAL
MAB_0190c|M.abscessus_ATCC_199 KAASAQHYLPNEGAELSFPMLQPTMLGGLCLIGTVWLVARARSSVRAGAL
.:.:****** ** * ***:. :: * :*: ** **: : *. *.**
Mflv_1176|M.gilvum_PYR-GCK AVAVLAVYAWSLLSMLTTLVGTTLLSFRLQPTLTILLAAAGAFGFVEVAR
Mvan_5629|M.vanbaalenii_PYR-1 AVAVLAVYAWSLLSMVTTLVGTTLLSFRLQPTLTILLAAAGAFGFIEVTR
MSMEG_6386|M.smegmatis_MC2_155 AVAVVAVYAWSLLSMLTTLAGTTLLSFRLQPTLTVLLTTAGAFGFIEATL
TH_1348|M.thermoresistible__bu GIAVLAVYAWSLLSMLTTLAGTTLLSFRLQPTLTVLFTAAGVFGFIEGAR
MAV_0230|M.avium_104 AIGVLAVYLWSLLSMLTTLARTTLLSFRLQPTLSVLLVAAGAFGLVEAAL
MLBr_00107|M.leprae_Br4923 MISVLAVYLWSLLSILTTLARTTLLSFRLQPTLTVLLVTAGVFGFIETAQ
Mb3821|M.bovis_AF2122/97 AIGVLAVYLWSLLSMLATLARTTLLSFRLQPTLSVLLVAAGAFGFVEAVQ
Rv3792|M.tuberculosis_H37Rv AIGVLAVYLWSLLSMLATLARTTLLSFRLQPTLSVLLVAAGAFGFVEAVQ
MMAR_5354|M.marinum_M AIGVLAVYLWSLLSMLTTLARTTLLSFRLQPTLTVLLATAGVFGFLEGTR
MUL_4971|M.ulcerans_Agy99 AIGVLAVYLWSLLSMLTTLARTTLLSFRLQPTLTVLLATAGIFGFLEGTR
MAB_0190c|M.abscessus_ATCC_199 AIGVTSVYLWSLLSMLATLAKTTLLSFRLVPTLTVLLAAAGVFAFLELTA
:.* :** *****:::**. ******** ***::*:.:** *.::* .
Mflv_1176|M.gilvum_PYR-GCK MAAARVRPPNARRVVAVATAVGAIGAVTFAQDIPDVLRSDIVVAYTDTDG
Mvan_5629|M.vanbaalenii_PYR-1 AAAARVRPANAPKVVAVATAVGVLGAVTFSQDIPDVLRSDIVVAYTDTDG
MSMEG_6386|M.smegmatis_MC2_155 AIAHRYRPETSRRVVAAATAVGAIGAVTFSQDIPDVLRPDINVAYTDTDG
TH_1348|M.thermoresistible__bu ALADSFS---SRRLLAVAATIGSIGALTFSQDIPDVLRYDIVVAYTDTDG
MAV_0230|M.avium_104 ALRRRSR-----AVIPVAGAIGLAAAIAFSQDIPDVLRPDLTIAYTDTDG
MLBr_00107|M.leprae_Br4923 SLAKHNR-----AVLSVASAIGLAGAIAFSQDIPNVLRPDLTIAYTDTDG
Mb3821|M.bovis_AF2122/97 ALGKRGR-----GVIPMAAAIGLAGAIAFSQDIPDVLRPDLTIAYTDTDG
Rv3792|M.tuberculosis_H37Rv ALGKRGR-----GVIPMAAAIGLAGAIAFSQDIPDVLRPDLTIAYTDTDG
MMAR_5354|M.marinum_M ALTTRSR-----AALPVAAAIGLAGAIAFSQDIPDVLRPDLTIAYTDTDG
MUL_4971|M.ulcerans_Agy99 ALTVRSR-----AALPVAAAIGLAGAIAFSQDIPDVLRPDLTIAYTDTDG
MAB_0190c|M.abscessus_ATCC_199 VAAKRTK-----HATLVATAIGLAGALAFSQDIPDVLRSDIVIAYADTDG
* ::* .*::*:****:*** *: :**:****
Mflv_1176|M.gilvum_PYR-GCK YGERADRRPPGAEQYYREVDARILEVTGRPRNETVVLTADYSFLSYYPYY
Mvan_5629|M.vanbaalenii_PYR-1 YGQRADRRPPGAEQYYREVDARIREVTGRPRNETVVLTADYSFLSYYPYY
MSMEG_6386|M.smegmatis_MC2_155 TGQRADRRPPGAERYYREIDAKILEVTGVPRNQTVVLTADYSFLSFYPYY
TH_1348|M.thermoresistible__bu NGERADRRPAGAERYYSEIDARILEVTGRPRDETVVLTADYSFLSYYPYY
MAV_0230|M.avium_104 HGQRGDRRPPSAEKFYASIDDVITRETGRPRDQTVVMTADYSFLSFYPYW
MLBr_00107|M.leprae_Br4923 HGQRGDRRPPGSEKYYWAIDEAVLHITGKPRDQTVVLTADYSFLAYYPYW
Mb3821|M.bovis_AF2122/97 YGQRGDRRPPGSEKYYPAIDAAIRRVTGKRRDRTVVLTADYSFLSYYPYW
Rv3792|M.tuberculosis_H37Rv YGQRGDRRPPGSEKYYPAIDAAIRRVTGKRRDRTVVLTADYSFLSYYPYW
MMAR_5354|M.marinum_M YGERGDQRPPGSEKYYSTIDTAIRKVTGKPRDQVVVMTADYSFLSYYPYW
MUL_4971|M.ulcerans_Agy99 YGERGDQRPPGSEKYYSTIDTTIRKVTGKPRDQVVVMTADYSFLSYYPYW
MAB_0190c|M.abscessus_ATCC_199 TGQRADRRPPGPESHYIAIDKAITEITGKPRDETVVLTADYSFLAFYPYY
*:*.*:**...* .* :* : . ** *:..**:*******::***:
Mflv_1176|M.gilvum_PYR-GCK GFQGLTSHYANPLAEFDKRAGAIEGWSMLTDADAFIAALDAMPWEPPSVF
Mvan_5629|M.vanbaalenii_PYR-1 GFQGLTSHYANPLAQFDQRAGAIEGWSMLTDADQFIEALDAMPWEPPSVF
MSMEG_6386|M.smegmatis_MC2_155 GFQGLTSHYANPLAQFDKRAAAIEGWATLGSADDFVAALDELPWEPPTVF
TH_1348|M.thermoresistible__bu GFQGLTSHYANPLAQFEQRAAAIESWAVLEDADQFIAALDALPWEPPTVF
MAV_0230|M.avium_104 GFQGLTSHYANPLAQFDLRAAQIDKWSKLKTADELLHALDTCPWPPPTVF
MLBr_00107|M.leprae_Br4923 GFQGLTSHYANPLAQFDLRAAQIQQWSRLTTASELIHALDTLPWPPPTVF
Mb3821|M.bovis_AF2122/97 GFQGLTPHYANPLAQFDKRATQIDSWSGLSTADEFIAALDKLPWQPPTVF
Rv3792|M.tuberculosis_H37Rv GFQGLTPHYANPLAQFDKRATQIDSWSGLSTADEFIAALDKLPWQPPTVF
MMAR_5354|M.marinum_M GFQGLTSHYANPLAQFDKRAAQIESWAKLKTSDEFVQALDKLPWPAPTVF
MUL_4971|M.ulcerans_Agy99 GFQGLTSHYANPLAQFDKRAAQIESWGKIKTSDEFVQALDKLLWPAPTVF
MAB_0190c|M.abscessus_ATCC_199 GFQGLTPHYANPLAQFDKRAAAIKSWEDLHTADEMVRALDALPWRAPTVL
******.*******:*: ** *. * : :. :: *** * .*:*:
Mflv_1176|M.gilvum_PYR-GCK LMR----RGAND----TYTLRLAEDVYPNQPNVRRYHVSLDDALFDDPRF
Mvan_5629|M.vanbaalenii_PYR-1 LMR----RGAND----TYTLRLAEDVYPNQPNVRRYHVALDDALFDDPRF
MSMEG_6386|M.smegmatis_MC2_155 LMR----HGAND----TYTLRLASDVYPNQPNVRRYHVELDSAVFDDPRF
TH_1348|M.thermoresistible__bu LLRGAGSGGADD----TYTLRLAEDVYPNQPNVRRYHVRLDRALFDDPRF
MAV_0230|M.avium_104 LMR----HGANN----TYTLRLAEDVYPNQPNVRRYTVELRAALFDSPRF
MLBr_00107|M.leprae_Br4923 VMR----HGAGN----TYTLRLAKNVYPNQPNVRRYTVDLPAALFADQRF
Mb3821|M.bovis_AF2122/97 LMR----HGAHN----SYTLRLAQDVYPNQPNVRRYTVDLRTALFADPRF
Rv3792|M.tuberculosis_H37Rv LMR----HGAHN----SYTLRLAQDVYPNQPNVRRYTVDLRTALFADPRF
MMAR_5354|M.marinum_M LMR----RGAGPGSGASYTLRLAEDVYPNQPNVRRYTVDLKAALFADPRF
MUL_4971|M.ulcerans_Agy99 LMR----RGAGPGSGASYTLRLAEDVYPNQPNVRRYTVDLKAALFADPRF
MAB_0190c|M.abscessus_ATCC_199 LMR----QSGDN-----YSLRLAKDVYPNQPNVKRYTVTLNKQLFADPRF
::* .. *:****.:********:** * * :* . **
Mflv_1176|M.gilvum_PYR-GCK DVSTIGPFVLAIRKPITI------
Mvan_5629|M.vanbaalenii_PYR-1 DVSTIGPFVLAIRKPSTI------
MSMEG_6386|M.smegmatis_MC2_155 EVSDHGPFVLAIRKPGGKPETDGH
TH_1348|M.thermoresistible__bu DVSTIGPFVLAIRVG---------
MAV_0230|M.avium_104 AVHRVGPFVLAVRKPAA-------
MLBr_00107|M.leprae_Br4923 AVQDIGPFVLAIRKPMGNA-----
Mb3821|M.bovis_AF2122/97 VVEDIGPFVLAIRKPQESA-----
Rv3792|M.tuberculosis_H37Rv VVEDIGPFVLAIRKPQESA-----
MMAR_5354|M.marinum_M TVETIGPFVLAIRKPAASA-----
MUL_4971|M.ulcerans_Agy99 TVETIGPFVLAIRKPAASA-----
MAB_0190c|M.abscessus_ATCC_199 TVRTIGPFVLVVRVPPHA------
* *****.:*