For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MATSDDAAAVAAVYLPYVRDTAISFETQQPSVEEMRSRLTTTLATLPWLVITDGPRVKGYAYASPHRARE AYRWSVDVSLYLDASIHRQGQGRRLYTALLNLLGAQGYINAYAAITLPNAASVGLHEALAFRRVGVFPRV GFKQQRWWDVGWWHRRLADPPAVPEEPRPWTRLPAPTINSALAVHPAQ
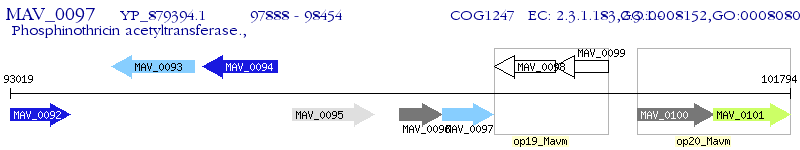
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0097 | - | - | 100% (188) | phosphinothricin N-acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3039c | - | 3e-35 | 46.98% (149) | phosphinothricin N-acetyltransferase |
| M. marinum M | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_0097|M.avium_104 ----MATSDDAAAVAAVYLPYVRDTAISFETQQPSVEEMRSRLTTTLATL
MAB_3039c|M.abscessus_ATCC_199 MHIRDATAADAAACAAIYAPYVTDTAISFEEVPPSDAVLAERIQTAAHRH
**: **** **:* *** ******* ** : .*: *:
MAV_0097|M.avium_104 PWLVITDGPRVKGYAYASPHRAREAYRWSVDVSLYLDASIHRQGQGRRLY
MAB_3039c|M.abscessus_ATCC_199 AWLVAEDGGEILGYAYAAPWKSRTAYQWACEVSVYVDGARRQRGTGRQLY
.*** ** .: *****:* ::* **:*: :**:*:*.: :::* **:**
MAV_0097|M.avium_104 TALLNLLGAQGYINAYAAITLPNAASVGLHEALAFRRVGVFPRVGFKQQR
MAB_3039c|M.abscessus_ATCC_199 TALLGRLRELGYRTVLAVVVTPNAASEKLHRTMGFEQVALYRRIGYKLGS
****. * ** .. *.:. ***** **.::.*.:*.:: *:*:*
MAV_0097|M.avium_104 WWDVGWWHRRLADPPAVPEEPRPWTRLPAPTINSALAVHPAQ
MAB_3039c|M.abscessus_ATCC_199 WHDVTHFQLFLTSAGDS--------RAPGPAPGEA-------
* ** :: *:.. * *.*: ..*