For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ATNSWVLPVLALTAALTVSCSHSRIQPQDASTSTSTSTSAVATNTAGRAKITFDPCSQIPASVIAQQKLD RLPPKPDRSADGDIENNRCGYLAQARYGVSAVASNYTLEMDKKVDFHGDFKEFDINGRRALSFLIYKNDP TACAIDVEATTGTYGVNASSAMGTFGDFPDCLTAARAHLDAFLPYFPA*
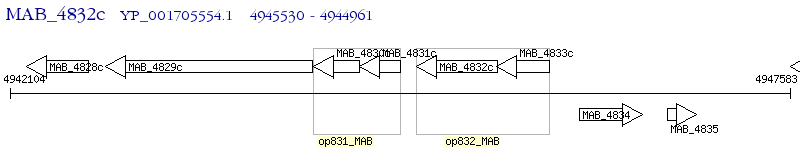
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4832c | - | - | 100% (189) | hypothetical protein MAB_4832c |
| M. abscessus ATCC 19977 | MAB_3120 | - | 2e-06 | 26.94% (193) | hypothetical protein MAB_3120 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2645 | - | 4e-05 | 27.40% (146) | hypothetical protein MSMEG_2645 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4832c|M.abscessus_ATCC_199 MATNSWVLPVL-ALTAALTVSCSHSRIQPQDASTSTSTSTSAVATNTAG-
MSMEG_2645|M.smegmatis_MC2_155 MAARVRLLSALCALVAAVILVWQTDPVAGTGSAAPRLDATDLPMTNVSTT
**:. :*..* **.**: : . . : .:::. :*. **.:
MAB_4832c|M.abscessus_ATCC_199 --------RAKITFDPCSQIPASVIAQQKLDRLPPKPDRSADGDIENNRC
MSMEG_2645|M.smegmatis_MC2_155 IKWPVIETTDPKPFDPCNDIPIDVIERIGLAWTPPMPE-------DGLRC
.****.:** .** : * ** *: :. **
MAB_4832c|M.abscessus_ATCC_199 GYLAQARYGVSAVASNYTLEMDKKVDFHGDFKEFDINGRRALSFLIYK--
MSMEG_2645|M.smegmatis_MC2_155 HYDAGN----YQMAVEAFVWRTYEQTIPADALELDIDGHRAAQYWVMKPT
* * :* : : : : .* *:**:*:** .: : *
MAB_4832c|M.abscessus_ATCC_199 --ND--PTACAIDVEATTGTYGVNASSAMGTFGDFPDCLTAARAHLDAFL
MSMEG_2645|M.smegmatis_MC2_155 DWNDRWWTTCMIAFDTSYGVIQQSLFYSPIHSPDKPDCLQTNLQRAHELA
** *:* * .::: *. . : * **** : : . :
MAB_4832c|M.abscessus_ATCC_199 PYFPA
MSMEG_2645|M.smegmatis_MC2_155 PYYKF
**: