For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQDSSGNQSLRHTSRPVTRRDALRYATAVSALAGLGAASLGTGAPTASAAAPTLIDFAMRQIPAEHIKAA GHAGVINYVSTSRPGSSMGAKPITRPYAQSLTAAGLVIVSNFQYGKPGGTAPSDFTRGFAGGVEDARTAW QLHTAAGGGQSAPIFFSVDDDIDRGTWNDVALQWFRGINSVLGVQRTGIYGGVNPCQWAASDGVIGNSRS PGHVWAWQTRSWSRGQVFPGAVLYQRIVSTASNPGPVVGGLEVDVSDALAQDVGQWNFHQ
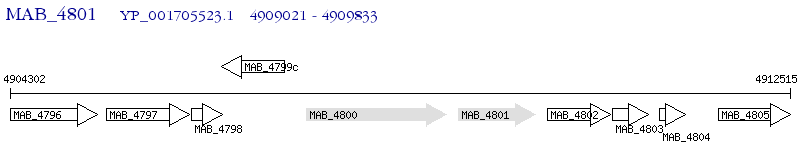
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4801 | - | - | 100% (270) | twin-arginine translocation pathway |
| M. abscessus ATCC 19977 | MAB_4782 | - | 5e-80 | 58.53% (258) | hypothetical protein MAB_4782 |
| M. abscessus ATCC 19977 | MAB_1511 | - | 8e-40 | 41.18% (255) | hypothetical protein MAB_1511 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2554c | - | 2e-38 | 40.16% (254) | hypothetical protein Mb2554c |
| M. gilvum PYR-GCK | Mflv_0504 | - | 1e-121 | 80.24% (253) | twin-arginine translocation pathway signal |
| M. tuberculosis H37Rv | Rv2525c | - | 2e-38 | 40.16% (254) | hypothetical protein Rv2525c |
| M. leprae Br4923 | MLBr_01190 | - | 5e-35 | 38.98% (254) | hypothetical protein MLBr_01190 |
| M. marinum M | MMAR_2953 | - | 1e-113 | 75.19% (258) | hypothetical protein MMAR_2953 |
| M. avium 104 | MAV_2754 | - | 1e-91 | 61.99% (271) | hypothetical protein MAV_2754 |
| M. smegmatis MC2 155 | MSMEG_6815 | - | 1e-116 | 76.95% (256) | secreted protein |
| M. thermoresistible (build 8) | TH_1407 | - | 1e-118 | 78.91% (256) | secreted protein |
| M. ulcerans Agy99 | MUL_2220 | - | 1e-112 | 74.42% (258) | hypothetical protein MUL_2220 |
| M. vanbaalenii PYR-1 | Mvan_5923 | - | 1e-121 | 81.03% (253) | twin-arginine translocation pathway signal |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0504|M.gilvum_PYR-GCK --------------MSITRRDALRYAA--AASALA-GLNAAAAALAPAAS
Mvan_5923|M.vanbaalenii_PYR-1 ----------------MSRRDALRYAT--AASALA-VLSAATGARTPTAS
TH_1407|M.thermoresistible__bu MRDAPEESDL-SMSRSISRRDALRFAA--GAALAG-MGAAAAGSGAPSAS
MAB_4801|M.abscessus_ATCC_1997 MQDSSGNQSLRHTSRPVTRRDALRYAT--AVSALAGLGAASLGTGAPTAS
MSMEG_6815|M.smegmatis_MC2_155 MKHSAE------RSRGISRRDALRYTA--AASVLA---GLGAGVSAAPAS
MMAR_2953|M.marinum_M -MHNLPLPPSAVRSRAISRRDALRYTT--ALAGLG---AASAACGMPTAA
MUL_2220|M.ulcerans_Agy99 -MHNLPLPPSAVRSRAISRRDALRYTT--ALAGLG---AASAACGMPTAA
MAV_2754|M.avium_104 -MRDSAGPGARGDRRPPSRREVLGYAAGCALAPVLLSLGTMAGAGGPTAR
Mb2554c|M.bovis_AF2122/97 --------------MSVSRRDVLKFAA-ATPGVLGLGVVASSLRAAPAS-
Rv2525c|M.tuberculosis_H37Rv --------------MSVSRRDVLKFAA-ATPGVLGLGVVASSLRAAPAS-
MLBr_01190|M.leprae_Br4923 --------------MSVSRRDVLKFAT-VTPGLLGLGVAAAALCAVPAST
:**:.* ::: . ..:
Mflv_0504|M.gilvum_PYR-GCK AAAP-TLIDFAMRQIPAQDIRAAGHSGVINYVSTSRPGSNFGA-KPITLP
Mvan_5923|M.vanbaalenii_PYR-1 AAAP-TLIDYAMRQIPAQDIRAAGHSGVINYVSTSRPGSSFGA-KPITLP
TH_1407|M.thermoresistible__bu AAPP-TLIDFAMRQIPPEAIRAAGHAGVINYVSTSRPGSNFGA-KPITLP
MAB_4801|M.abscessus_ATCC_1997 AAAP-TLIDFAMRQIPAEHIKAAGHAGVINYVSTSRPGSSMGA-KPITRP
MSMEG_6815|M.smegmatis_MC2_155 AAAP-QLIDFAAAQIPAEHIKAAGYAGVINYVSTSRPGSSFGA-KPITRP
MMAR_2953|M.marinum_M AAAPPRLIDFAAHQIPAQQIRAAGYSGVVNYVSLSRPGSSFGA-KPITRR
MUL_2220|M.ulcerans_Agy99 AAAPPRLIDFAAHQIPAQQIRAAGYSGVVNYVSLSRPGSSFGA-KPITRR
MAV_2754|M.avium_104 AAGL-KLIDFAERRIAPEEIKAAGYDGVVNYVSLSRPGADFEA-KPLTRD
Mb2554c|M.bovis_AF2122/97 AGSLGTLLDYAAGVIPASQIRAAGAVGAIRYVSDRRPGGAWMLGKPIQLS
Rv2525c|M.tuberculosis_H37Rv AGSLGTLLDYAAGVIPASQIRAAGAVGAIRYVSDRRPGGAWMLGKPIQLS
MLBr_01190|M.leprae_Br4923 AGSLGTLLDYAAGVIPASQIRATGAVGAIRYVSD--PRGTWAVGKPIQVT
*. *:*:* *... *:*:* *.:.*** * . **:
Mflv_0504|M.gilvum_PYR-GCK YAKQLTAAGLVIVSNYQYGKPGGTAPSDFTRGFAGGVADARTGWALHTAA
Mvan_5923|M.vanbaalenii_PYR-1 YAKSLVAAGLVIVSNYQYGKPGGTAPSDFTRGYAGGVADARTGWALHTAA
TH_1407|M.thermoresistible__bu YAKALTAAGLVIVSNYQYGKPGGTAPSDFTRGFAGGVADARTGWHHHTAA
MAB_4801|M.abscessus_ATCC_1997 YAQSLTAAGLVIVSNFQYGKPGGTAPSDFTRGFAGGVEDARTAWQLHTAA
MSMEG_6815|M.smegmatis_MC2_155 YAESLTRAGLVIVSNYQYGKPGGTAPSDFRRGAAGGVADAKTAWQLHTAA
MMAR_2953|M.marinum_M YADSLTAAGLVIVSNYQYGKPGGSAPSDFTRGYAGGVADARTAWQLHTAA
MUL_2220|M.ulcerans_Agy99 YADSLTAAGLVIVSNYQYGKPGGSAPSDCTRGYAGGVADARTAWQLHTAA
MAV_2754|M.avium_104 YADALRAAGLHIVSNFQYGKPGWPDPSDFTRGYDGGVADAHTATQLHAAA
Mb2554c|M.bovis_AF2122/97 EARDLSGNGLKIVSCYQYGK-GST--ADWLGGASAGVQHARRGSELHAAA
Rv2525c|M.tuberculosis_H37Rv EARDLSGNGLKIVSCYQYGK-GST--ADWLGGASAGVQHARRGSELHAAA
MLBr_01190|M.leprae_Br4923 EARDLINNGLKIVSCYQYGK-GNT--ADWLGGATAGLRHAQRGVQLHTAA
* * ** *** :**** * . :* * .*: .*: . *:**
Mflv_0504|M.gilvum_PYR-GCK GGGKSAPIFFSVDDDIDRQAWNDLALPWFRGINSVIGVQRTGIYAGIRPC
Mvan_5923|M.vanbaalenii_PYR-1 GGGQSAPIFFSVDDDIDRETWNRLALPWFRGINSVIGVQRTGIYAGIRPC
TH_1407|M.thermoresistible__bu GGGQSAPIYFSVDDDIDRHTWNTLALPWFRGINSVLGVQRTGVYGGINVV
MAB_4801|M.abscessus_ATCC_1997 GGGQSAPIFFSVDDDIDRGTWNDVALQWFRGINSVLGVQRTGIYGGVNPC
MSMEG_6815|M.smegmatis_MC2_155 GGARNAPIYFSVDEDIDRQTWNNLALPWFRGINSVIGVQRTGVYGGVNVC
MMAR_2953|M.marinum_M GGGQGAPVFFTVDEDINRDTWNRVALQWFRGINSVLGVQRTGVYGGIDVC
MUL_2220|M.ulcerans_Agy99 GGGQGAPVFFTVDEDINRDTWNRVALQWFRGINSVLGVQRTGVYGGIDVC
MAV_2754|M.avium_104 GGPDSAPIFFSVDDDIDADTWNNVAVQWFRGINSVLGVGRTGIYGHARAC
Mb2554c|M.bovis_AF2122/97 GGPTSAPIYASIDDNPSYEQYKNQIVPYLRSWESVIGHQRTGVYANSKTI
Rv2525c|M.tuberculosis_H37Rv GGPTSAPIYASIDDNPSYEQYKNQIVPYLRSWESVIGHQRTGVYANSKTI
MLBr_01190|M.leprae_Br4923 GGPVSAPIYASIDSNPTYEQYKQQVAPYLRSWESVIGHQRTGVYANSRTI
** .**:: ::*.: :: ::*. :**:* ***:*.
Mflv_0504|M.gilvum_PYR-GCK QWATADGVIGRSGTPGRVWAWQTRSWS-NGQIYPGAVLYQRIIDTASNPG
Mvan_5923|M.vanbaalenii_PYR-1 QWATADGVIGRSRTPGKVWAWQTRSWS-NGQIYPGAVLYQRIIDTASNPG
TH_1407|M.thermoresistible__bu QWAIEDGVIGFSRVQGRRWAWQTRSWS-RGQIHPAAVLYQRIIDTPSNPG
MAB_4801|M.abscessus_ATCC_1997 QWAASDGVIGNSRSPGHVWAWQTRSWS-RGQVFPGAVLYQRIVSTASNPG
MSMEG_6815|M.smegmatis_MC2_155 QWAIEDGVIGRSGTPGKAWAWQTRSWS-KGKIHPAAVLYQHTIDTASNPG
MMAR_2953|M.marinum_M QWAAADGVIGSSGTPGRRWAWQTRAWS-GHRIYPAAVLYQRVVSTKSSPG
MUL_2220|M.ulcerans_Agy99 QWAAADGVIGSSGTPGRRWAWQTRAWS-GHRIYPAAVLYQRVVSTKSSPG
MAV_2754|M.avium_104 GWAIRDGVVGPSSTPGHRWAWQTRSWS-HGEREPAAVLYQAVVNSPGSPG
Mb2554c|M.bovis_AF2122/97 DWAVNDGLGS--------YFWQHNWGSPKGYTHPAAHLHQVEIDKRK---
Rv2525c|M.tuberculosis_H37Rv DWAVNDGLGS--------YFWQHNWGSPKGYTHPAAHLHQVEIDKRK---
MLBr_01190|M.leprae_Br4923 AWALQDGLAS--------YFWQHNWGSPKGYTHPAANLHQVEIDRRT---
** **: . : ** . * *.* *:* :.
Mflv_0504|M.gilvum_PYR-GCK PIVGGIRVDVNDVLAQDCGQWNLHP--------
Mvan_5923|M.vanbaalenii_PYR-1 PVVGGIRVDVNDVLAQDCGQWNLHP--------
TH_1407|M.thermoresistible__bu PVVGGIRVDVNDVLAADVGQWNLHP--------
MAB_4801|M.abscessus_ATCC_1997 PVVGGLEVDVSDALAQDVGQWNFHQ--------
MSMEG_6815|M.smegmatis_MC2_155 PLVGGIRVDVNDALAQDIGQWNFHP--------
MMAR_2953|M.marinum_M PRVGGFEVDVNDVLAPDCGQWNLHQGHRNGDAR
MUL_2220|M.ulcerans_Agy99 PRVGGFEVDINDVLAPDCGQWNLHQGHRNGDAR
MAV_2754|M.avium_104 PLLGGINVDVDDVLAADYGQWDFAR--------
Mb2554c|M.bovis_AF2122/97 --VGGVGVDVNQILKPQFGQWA-----------
Rv2525c|M.tuberculosis_H37Rv --VGGVGVDVNQILKPQFGQWA-----------
MLBr_01190|M.leprae_Br4923 --VGGVGVDVNTILKPQFGQWA-----------
:**. **:. * : ***