For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPTTASTPAVVGGLRRQLLRSICTTALFMAPALITRVGGLHPSPVLALVIYGAAVVAASFVLAWAAEAAQ IDVSGGLAIAVLALIAVLPEYAVDLYYAYISGHVPEYTQYAAANMTGSNRLLMGLGWPVVVLLALAVARK TSGHPVNLLSLEPSNRIELGFLTVAGVVAFVIPASGQIHLALGIALLAWFGFYLFTISRGEAEEPDLVGT AAAIGMLPKHLRRSTVVTMFVLAASVILLCAEPFANSLVSAGTQLGVDQFLLVQWLAPLASEAPEFIIAA IFAARGKGTAAIATLISSKVNQWTLLIGSLPLAHLAGGGGFSLVLDARQVEETLLTATQTMMGVALILAL RFHRATAWALLALFVVQFPITSTHGRLILCGVYGVLAIAGLIVNRHHILATIRSPFHRSATLQLTVP
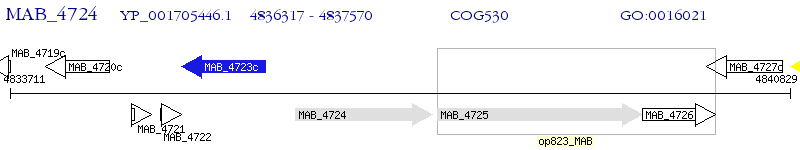
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4724 | - | - | 100% (417) | Sodium/calcium exchanger family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5095 | - | 2e-07 | 24.74% (291) | CaCA family Na(+)/Ca(+) antiporter |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_5239 | - | 1e-161 | 72.47% (396) | sodium/calcium exchanger family protein |
| M. smegmatis MC2 155 | MSMEG_4754 | - | 2e-07 | 21.60% (338) | K+-dependent Na+/Ca+ exchanger related-protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5095|M.gilvum_PYR-GCK -----------------------MNG--LWFVVGLITVIVG-------AE
MSMEG_4754|M.smegmatis_MC2_155 -----------------------MLGSAAWFVAGLLALAFG-------SE
MAB_4724|M.abscessus_ATCC_1997 ---MPTTASTPAVVGGLRRQLLRSICTTALFMAPALITRVGGLHPSPVLA
MAV_5239|M.avium_104 MLAIDRPAAVATTARWRRRTLTRSGLITGAFVAPAVAVRITGMHTGAVAA
*:. : .
Mflv_5095|M.gilvum_PYR-GCK VMVRGGAEVAARLGISPIIIGLTVVSIG----------TSMPELAVGVVA
MSMEG_4754|M.smegmatis_MC2_155 VMVRGGAQMASRMGISPIVVGMTVVSIG----------TSLPELAVGVTA
MAB_4724|M.abscessus_ATCC_1997 LVIYGAAVVAASFVLAWAAEAAQIDVSGGLAIAVLALIAVLPEYAVDLYY
MAV_5239|M.avium_104 LLIFGAAVVAASFLLAWAAETAQIDVSGGLATALLALIAVLPEYAVDLYY
::: *.* :*: : :: : * : :** **.:
Mflv_5095|M.gilvum_PYR-GCK AADGSGALAVGNIAGTNVVN---ILLILGLS-ALLVPLGLAMRTIRFELP
MSMEG_4754|M.smegmatis_MC2_155 ATEGSGALAVGNIAGTNTVN---LLFILGLS-ALLRPLAIQRRTLRLDLP
MAB_4724|M.abscessus_ATCC_1997 AYISGHVPEYTQYAAANMTGSNRLLMGLGWPVVVLLALAVARKTSGHPVN
MAV_5239|M.avium_104 AYVSGHNADYTQYAAANMTGSNRLLMGLGWPVVVLVSIVVARRSGSDRPA
* .. : *.:* .. :*: ** . .:* .: : ::
Mflv_5095|M.gilvum_PYR-GCK MMAVAALLFWGLAADGNLSRLDGGILVTAAVGYTAAVIWSARRESRAIAA
MSMEG_4754|M.smegmatis_MC2_155 VMAVAAVLFWVLCIDGTLSRVDGVILLCAAVAYTVVVVRSARVESRDVVD
MAB_4724|M.abscessus_ATCC_1997 LLSLEPSNRIELGFLTVAGVVAFVIPASGQIHLALGIALLAWFGFYLFTI
MAV_5239|M.avium_104 GLALLPANRVELGFLLIAGVIAFAIPASGQIHFGLGLALLAWFGFYLYKI
::: . * . : * . : : *
Mflv_5095|M.gilvum_PYR-GCK EFASEFAPEPAATTTEVRRRT--------ATHVAMTVGGIVVVVIGADWL
MSMEG_4754|M.smegmatis_MC2_155 EYAYAYRGEPAGDDGAAEREPDMPDTGSLVRYFAMTIVGITVVVVGADWL
MAB_4724|M.abscessus_ATCC_1997 SRGEAEEPDLVGTAAAIGMLPKHLRR---STVVTMFVLAASVILLCAEPF
MAV_5239|M.avium_104 SHGNVEEPDLIGTAAALGELPDGRRR---IAVVGLFLVSGAVILLCAKPF
. . : . . . : : . *::: *. :
Mflv_5095|M.gilvum_PYR-GCK VDGAVGMARQFGVTDALIGLTVVAIGTSAPELVTTVMTTLRGDRDIAIGN
MSMEG_4754|M.smegmatis_MC2_155 VEGAVGIAREFGVSDALIGLTIVAIGTSAPELVTTIVSTVRGERDIAIGN
MAB_4724|M.abscessus_ATCC_1997 ANSLVSAGTQLGVDQFLLVQWLAPLASEAPEFIIAAIFAARGKGTAAIAT
MAV_5239|M.avium_104 ADNLVAAGTELGVNRFLLVQWLAPLASEAPEFIIATIFASRGKGTTAIAT
.:. *. . ::** *: :..:.:.***:: : : : **. **..
Mflv_5095|M.gilvum_PYR-GCK LIGSSIYNILLILG---ITCLIPAGGLALPDSLVRLDIPIMVAVAGLCIP
MSMEG_4754|M.smegmatis_MC2_155 LLGSSVYNIVLILG---ATCLVPADGLPLNSSLVWIDIPVMVVATLLLIP
MAB_4724|M.abscessus_ATCC_1997 LISSKVNQWTLLIGSLPLAHLAGGGGFSLVLDARQVEETLLTATQTMMGV
MAV_5239|M.avium_104 LISSKVNQWTLLIGSLPLAHLLGGGGFALHLDSRQVEEVLLTATQTMMGV
*:.*.: : *::* : * ..*:.* . :: ::... :
Mflv_5095|M.gilvum_PYR-GCK IFLSGRRVSRMEGGLMVAAYLGYLAFVLSTQT------------------
MSMEG_4754|M.smegmatis_MC2_155 VFFSGRRVHRAEGGAMVVAYLAYLTFLLFTQT------------------
MAB_4724|M.abscessus_ATCC_1997 ALILALRFHRATAWALLALFVVQFPITSTHGRLILCGVYGVLAIAGLIVN
MAV_5239|M.avium_104 ALILALRFHRSAALALLALFVVQFPIVSTQGRLLLCGVYTAVAVVALIGY
:: . *. * . ::. :: :.:
Mflv_5095|M.gilvum_PYR-GCK ------------------------------
MSMEG_4754|M.smegmatis_MC2_155 ------------------------------
MAB_4724|M.abscessus_ATCC_1997 RHHILATIRSPFHRSATLQLTVP-------
MAV_5239|M.avium_104 RRHLAATLRAPFFGTAIRHSGHPHHPVPDP