For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSHTVPTAQASLSERVDAPDVVEIPSAGADLTWRAATKEDIPALFELWRAAGAVDHPTSLVMLDELEEEF DDDDFDPALDSVIAVDSLGRVVAFGSATVKSAHETVVWVALDGTVHPERRGEGIGSSVLRWQEQRGLQHL AESDECLPGWLASSAEEHAVWTIELFHRNGYESVRWWHELERDLAQPIPDVTLPEGIRIETYGPEWSEPT RDAHNEAFRDHWGSQPEAREDWEAAHRLSAFRADLSFVAVARDAAGQDIVVAYLLSDVNEEEWEANGYSF GFVDLLGVRRDWRGRKLAQALLTHAMRAYRHEGLQRAVLDVDADSPTGAVALYEGLGFSLVNRSISLIKQ F*
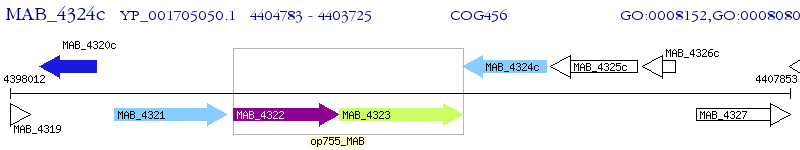
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4324c | - | - | 100% (352) | putative acetyltransferase, GNAT |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1651 | - | 2e-05 | 27.50% (240) | GCN5-related N-acetyltransferase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4548 | - | 6e-10 | 30.15% (136) | acetyltransferase, gnat family protein |
| M. thermoresistible (build 8) | TH_0384 | - | 4e-05 | 27.27% (231) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1651|M.gilvum_PYR-GCK -------------------------------MSWQQSLTDDRARQIRELV
TH_0384|M.thermoresistible__bu --------------------------------------------------
MAB_4324c|M.abscessus_ATCC_199 MTSHTVPTAQASLSERVDAPDVVEIPSAGADLTWRAATKEDIPALFELWR
MSMEG_4548|M.smegmatis_MC2_155 ----------------------MTQNIGVGTWTVRQATSHDHPHIADFLA
Mflv_1651|M.gilvum_PYR-GCK SAASAADGVAPVGDQVLRALAHDR-----TRHLVAVEDDGAGSRVRGYLN
TH_0384|M.thermoresistible__bu --------------------------------------------------
MAB_4324c|M.abscessus_ATCC_199 AAGAVDHPTSLVMLDELEEEFDDDDFDPALDSVIAVDSLGRVVAFGSATV
MSMEG_4548|M.smegmatis_MC2_155 TTTGLGGRTFAADSRDVAEQLDGALPG---SAVVLRGESGGVLGYAALHG
Mflv_1651|M.gilvum_PYR-GCK LAPADDDAP--PMAEVVVHPDARRRGTGSAMIRAA-------LAEGGPQT
TH_0384|M.thermoresistible__bu ------------MAELVVAPQARRRGIGSEMIRRG-------LAAGGADA
MAB_4324c|M.abscessus_ATCC_199 KSAHETVVW--VALDGTVHPERRGEGIGSSVLRWQEQRGLQHLAESDECL
MSMEG_4548|M.smegmatis_MC2_155 PDTEEPEVLGTFVFAPSAPIQAVRDIVGDSVDHFHR------VATPGSYL
. : *. : : :* .
Mflv_1651|M.gilvum_PYR-GCK RVWAHGNLVAARATAEALGLT----AVRELLQMQRPLAD-LPPQTTAQRV
TH_0384|M.thermoresistible__bu RVWAHGNLEPAQAVAAVLGMR----VVRELLQMRRPLTG-LPPVQVPDGV
MAB_4324c|M.abscessus_ATCC_199 PGWLASSAEEHAVWTIELFHRNGYESVRWWHELERDLAQPIPDVTLPEGI
MSMEG_4548|M.smegmatis_MC2_155 RVYIGADQPAAIAALTETGAR----RERQFISTRRSLLGEDSDQLAAEHI
: .. . * . .* * . .: :
Mflv_1651|M.gilvum_PYR-GCK R------FATYSGPHDDAEVLRVNNAAFSWHPEQGGWTDADIAERRGEPW
TH_0384|M.thermoresistible__bu R------IFTYPGPDADAELLRVNNAAFAWHPEQGDLTAEDLAEQRAEPW
MAB_4324c|M.abscessus_ATCC_199 R------IETY-GPEWSEPTRDAHNEAFRDHWGSQPEAREDWEAAHRLSA
MSMEG_4548|M.smegmatis_MC2_155 DGLTVLSWPQVISGGFGEEVRRLQFDTFSEHFGNMSKTPQRWEHHLHSRA
. . : :* * . :
Mflv_1651|M.gilvum_PYR-GCK FDPAGFFLAFDEDTG---ALLGFHWTKVHSE-------SLGEVYVVGVDP
TH_0384|M.thermoresistible__bu FDPDGLFLAADADTS---KLLGFHWTKVHSD-------RLGEVYVVGVDP
MAB_4324c|M.abscessus_ATCC_199 FRADLSFVAVARDAAGQDIVVAYLLSDVNEEEWEANGYSFGFVDLLGVRR
MSMEG_4548|M.smegmatis_MC2_155 FTPDFSLAAVDRDGVVVGYVLGSTYSVLTDRGEE----RSAHTDYIGVRR
* . : * * ::. : : . . . :**
Mflv_1651|M.gilvum_PYR-GCK AAQGRGLGAALTLLGLHHLESRLTGDGRVADVMLYVEADN-TAAVKTYRR
TH_0384|M.thermoresistible__bu AAQGRGLGRVLTLIGLHHLAERLS-DSSHPMVMLYVEADN-AAAVATYRK
MAB_4324c|M.abscessus_ATCC_199 DWRGRKLAQALLTHAMRAYR-----HEGLQRAVLDVDADSPTGAVALYEG
MSMEG_4548|M.smegmatis_MC2_155 DRRERGTGELLLKKIWLAALR-----RGLTAASLGTDINNASNAHVLYRR
: * . * . * .: :. : * *.
Mflv_1651|M.gilvum_PYR-GCK LGFDIVNTDVAYAAVAPTDV----
TH_0384|M.thermoresistible__bu LGFEVFGVDVAYAFTTPGDTPAGQ
MAB_4324c|M.abscessus_ATCC_199 LGFSLVNRSISLIKQF--------
MSMEG_4548|M.smegmatis_MC2_155 LGYVAVRDEYAYRIDADGSTK---
**: . . :