For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSTGGQDREEEREPVTVTDKRRIDPNTGAVREEAKAAESKAATAEPGASASGAAGSATSGPAAAASEGGS DADVKVAELTADLQRAHADFANYRKRVERDRQAVIDSAKASVVTQLLGVLDDLDRAREHGDLESGPLRSV SDKLTAALEGLGLATFGAEGDDFDPSLHEAVQHDGQDGHPVLAAVLRKGYKLGDRVLRTAMVVVTDGDTA QQEPGTGDAETKSETESE*
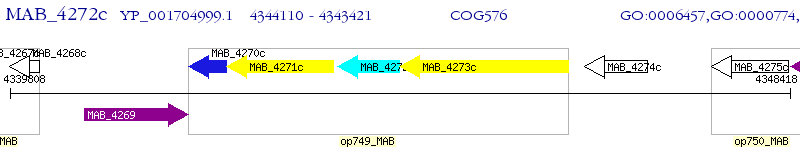
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4272c | - | - | 100% (229) | protein GrpE (HSP-70 cofactor) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0359 | grpE | 2e-56 | 54.67% (225) | GRPE protein (HSP-70 cofactor) |
| M. gilvum PYR-GCK | Mflv_0259 | - | 5e-60 | 54.26% (223) | GrpE protein |
| M. tuberculosis H37Rv | Rv0351 | grpE | 3e-56 | 54.67% (225) | GRPE protein (HSP-70 cofactor) |
| M. leprae Br4923 | MLBr_02495 | grpE | 7e-56 | 54.71% (223) | heat shock protein GrpE |
| M. marinum M | MMAR_0638 | grpE | 7e-53 | 50.68% (219) | GrpE protein (Hsp-70 cofactor) |
| M. avium 104 | MAV_4807 | - | 1e-60 | 57.59% (224) | protein GrpE |
| M. smegmatis MC2 155 | MSMEG_0710 | grpE | 4e-59 | 51.71% (234) | co-chaperone GrpE |
| M. thermoresistible (build 8) | TH_1022 | grpE | 5e-59 | 54.42% (226) | co-chaperone GrpE |
| M. ulcerans Agy99 | MUL_0594 | grpE | 2e-52 | 50.23% (219) | GrpE protein (Hsp-70 cofactor) |
| M. vanbaalenii PYR-1 | Mvan_0635 | - | 1e-58 | 53.15% (222) | GrpE protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0359|M.bovis_AF2122/97 MTDGNQKPDGNSGEQVTVTDKRRIDPETGEVRHVPP-----------GDM
Rv0351|M.tuberculosis_H37Rv MTDGNQKPDGNSGEQVTVTDKRRIDPETGEVRHVPP-----------GDM
MMAR_0638|M.marinum_M MSEGNPQ------EQVTVTDKRRVDPETGEVRHVPP-----------GDT
MUL_0594|M.ulcerans_Agy99 MSEGNPQ------EQVTVTDKRRVDPETGEVRHVPP-----------GDT
MLBr_02495|M.leprae_Br4923 MTEGNPH------EQVTVTDKRRIDPDTGEVRSIPL-----------GDT
MAV_4807|M.avium_104 MTQGNQKTEGNPPEQVTVTDKRRIDPETGEVRHVPP-----------GDT
Mflv_0259|M.gilvum_PYR-GCK MSDADRVE-----EPITVTDKRRIDPDTGQVREN-----------PSEPA
Mvan_0635|M.vanbaalenii_PYR-1 MSDPVDRE-----EPITVTDKRRIDPDTGQVREDRE-NS------DAGPA
TH_1022|M.thermoresistible__bu -VTGNDSH-----EPVTVTDKRRIDPVTGQKRDVDSGAA------PSGPA
MSMEG_0710|M.smegmatis_MC2_155 -MTQDDSH-----EPVTITDKRRIDPETGEVREP-------------AAT
MAB_4272c|M.abscessus_ATCC_199 MTSTGGQDREEEREPVTVTDKRRIDPNTGAVREEAKAAESKAATAEPGAS
* :*:*****:** ** *
Mb0359|M.bovis_AF2122/97 PGGTAAAD--------AAHTED---KVAELTADLQRVQADFANYRKRALR
Rv0351|M.tuberculosis_H37Rv PGGTAAAD--------AAHTED---KVAELTADLQRVQADFANYRKRALR
MMAR_0638|M.marinum_M PGGTPPAG--------ADSSAA---KVAELTADLQRVQADFANYRKRALR
MUL_0594|M.ulcerans_Agy99 PGGTPPAG--------ADSSAA---KVAELTADLQRVQADFANYRKRALR
MLBr_02495|M.leprae_Br4923 PGGSEPAGMTDRSANSTDTMVD---KVAELTSDLQRVQADFANYRKRALR
MAV_4807|M.avium_104 PGGTAPQAATAES---GGAAAD---KVAELTADLQRVQADFANYRKRALR
Mflv_0259|M.gilvum_PYR-GCK PSGPATDEFAG----ETEEEAG---KAAELLADLQRVQADFSNYRRRTLR
Mvan_0635|M.vanbaalenii_PYR-1 PSGPAPDEFAG----ESVEEAG---KAAELLADLQRVQADFANYRKRALR
TH_1022|M.thermoresistible__bu PGSAPADPSAAKA--EQAEKSGEAERAAELLADLQRVQADFANYRKRTLR
MSMEG_0710|M.smegmatis_MC2_155 PQGSAPASAAP----ETGGDSD---EVTELKATLQRVKAEYDNYRKRALR
MAB_4272c|M.abscessus_ATCC_199 ASGAAGSATSGPAAAASEGGSDADVKVAELTADLQRAHADFANYRKRVER
. .. ..:** : ***.:*:: ***:*. *
Mb0359|M.bovis_AF2122/97 DQQAAADRAKASVVSQLLGVLDDLERARKHGDLESGPLKSVADKLDSALT
Rv0351|M.tuberculosis_H37Rv DQQAAADRAKASVVSQLLGVLDDLERARKHGDLESGPLKSVADKLDSALT
MMAR_0638|M.marinum_M DQQAAADRAKASVVSELLHAVDDIERARKHGDLDSGPLKAVADKMMSVLT
MUL_0594|M.ulcerans_Agy99 DQQAAADRAKASVVSELLHAVDDIERARKHGDLDFGPLKAVADKMMSVLT
MLBr_02495|M.leprae_Br4923 DQQTASDRAKATVISQLLGVLDDFDRAREHGDLDSGPLKSVADKLMSALT
MAV_4807|M.avium_104 DQQAAADRAKAAVVNQLLGVLDDLERARKHGDLESGPLKSVADKLESALT
Mflv_0259|M.gilvum_PYR-GCK DQQVIADRAKASVITQLLPVLDDLDRARSHGDLDSGPLKAVADKIVTTLE
Mvan_0635|M.vanbaalenii_PYR-1 DQQLMADRAKATVVSQLLPVLDDLDRARSHGDLESGPFKAVADKLVAILE
TH_1022|M.thermoresistible__bu DQQAVAERTKASVIGELLPILDDLDHARRHGDLEAGPLKAFADKLVNTLE
MSMEG_0710|M.smegmatis_MC2_155 DQQLIAERTKANVVSELLGVLDDLDRARSHGDLESGPLKAVADKLVSTLE
MAB_4272c|M.abscessus_ATCC_199 DRQAVIDSAKASVVTQLLGVLDDLDRAREHGDLESGPLRSVSDKLTAALE
*:* : :** *: :** :**:::** ****: **:::.:**: *
Mb0359|M.bovis_AF2122/97 GLGLVAFGAEGEDFDPVLHEAVQHEGDGGQGSKPVIGTVMRQGYQLGEQV
Rv0351|M.tuberculosis_H37Rv GLGLVAFGAEGEDFDPVLHEAVQHEGDGGQGSKPVIGTVMRQGYQLGEQV
MMAR_0638|M.marinum_M GLGLKSFGAEGEDFDPVLHEAVQHEGDGGQDAKPVIGTVMRQGYQLGEHV
MUL_0594|M.ulcerans_Agy99 GLGLKSFGAEGEDFDPVLHEAVQHEGDGGQDAKPVIGTVMRQGYQLGEHV
MLBr_02495|M.leprae_Br4923 GLGLVAFGVEGEDFDPVLHEAVQHEGDGGEGSKPVIGDVLRHGYKLGDQV
MAV_4807|M.avium_104 GLGLTAFGEEGEEFDPVLHEAVQHEGDG---SKPVIGTVMRQGYKLGDQV
Mflv_0259|M.gilvum_PYR-GCK GLGLSGFGEEGDEFDPELHEAVQHEGEG---THPVLGSVMRRGYKVGDVV
Mvan_0635|M.vanbaalenii_PYR-1 GFGLSGFGEEGDEFDPALHEAVQHEGEG---THPVVGTVMRRGYRVGEQV
TH_1022|M.thermoresistible__bu SLGLSSFGAEGEDFDPELHEAVQHEGNG---GHPIIGTVLRRGYKVGDMV
MSMEG_0710|M.smegmatis_MC2_155 GLGLSAFGEEGDEFDPQLHEAVQHEGDG---THPVVGTVMRRGYRVGEQV
MAB_4272c|M.abscessus_ATCC_199 GLGLATFGAEGDDFDPSLHEAVQHDGQD---GHPVLAAVLRKGYKLGDRV
.:** ** **::*** *******:*:. :*::. *:*:**::*: *
Mb0359|M.bovis_AF2122/97 LRHALVGVVDTVVVDAAELESVDDGTAVADTAENDQADQGNSADTLGEQA
Rv0351|M.tuberculosis_H37Rv LRHALVGVVDTVVVDAAELESVDDGTAVADTAENDQADQGNSADTSGEQA
MMAR_0638|M.marinum_M LRNALVAVVETIADDTSEAGSTQQ--PADS-GEAGRPDTADNADAPAE--
MUL_0594|M.ulcerans_Agy99 LRNALVAVVETIADDTSEAGSTQQ--PADS-GEAGRPDTADNADAPAE--
MLBr_02495|M.leprae_Br4923 LRHALVGVVDTIAGDGAETVAIVA--PVDSTAKTEQGELGDNVTPHKEDG
MAV_4807|M.avium_104 LRHALVGVVDTVTEEGDGEAAATD-EPTAAAAETRPPESDDNAGASGD--
Mflv_0259|M.gilvum_PYR-GCK VRHAMVGVVDTVPDASGSGNADSG--AQQ-------PSES----------
Mvan_0635|M.vanbaalenii_PYR-1 VRHALVGVVDTIPDASGPANTGSE--PEQ-------PAES----------
TH_1022|M.thermoresistible__bu IRHAMVAVVDTEPGGDTPDNDGAA--DGQ-------AAESDTTESNND--
MSMEG_0710|M.smegmatis_MC2_155 IRHAMVGVVDTVPDNASEDGSGAQ--PEDGSGDKPDAAAEQAAESDN---
MAB_4272c|M.abscessus_ATCC_199 LRTAMVVVTDG---DTAQQEPGTG-------------DAETKSETESE--
:* *:* *.:
Mb0359|M.bovis_AF2122/97 ESEPSGS
Rv0351|M.tuberculosis_H37Rv ESEPSGS
MMAR_0638|M.marinum_M -------
MUL_0594|M.ulcerans_Agy99 -------
MLBr_02495|M.leprae_Br4923 D------
MAV_4807|M.avium_104 -------
Mflv_0259|M.gilvum_PYR-GCK -------
Mvan_0635|M.vanbaalenii_PYR-1 -------
TH_1022|M.thermoresistible__bu -------
MSMEG_0710|M.smegmatis_MC2_155 -------
MAB_4272c|M.abscessus_ATCC_199 -------