For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSAAQILSLIMLVVVLAAAIWRRMNIGVLALGAALPVLALSGLDAKAMYTTFPGDIFALIVGVSLLFAHL ERSGTLSWFVELVYGAVGDRHVLLPWAGFLVGAILSTAGAFSTAVIAFLVPLVAHVAVRYRSSFYLCELG VVIGANSAGLSPLNPTGAVVRAAATKFHVDYPGWALWAVSMVVAALVVLCLQLLDAFRRRRGGGFDLTPA PAAATETDAAESPSLRYAWCAGLALLTFVLLVVVVKTDVGVTAIALAALMQIAFHPHERSLLARIPWPSI LLLCGLLTYLGLMQKIGTIDSIASVLHRLGTGAVLILVLAYLTALLCNIESSTLGVLGLMMPIVFSTFTE PAQMFWVVCAVCVPAALMVMNPIHVAGTLIIGNSAADEQDNLFRRLLALAGSLALVVPGLLSIVPIALA
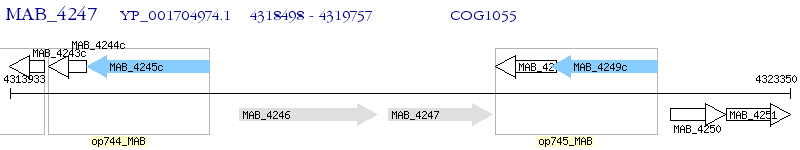
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4247 | - | - | 100% (419) | putative dicarboxylate carrier protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4611 | - | 1e-41 | 27.59% (424) | carrier protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4247|M.abscessus_ATCC_1997 MSAAQILSLIMLVVVLAAAIWRRMNIGVLALGAALPV-LALSGLDAKAMY
MSMEG_4611|M.smegmatis_MC2_155 -MSVQLLSIAVLVVVFVISTVFNVHMGALAFVATFVVGIGLTDLSEKEII
:.*:**: :****:. : .:::*.**: *:: * :.*:.*. * :
MAB_4247|M.abscessus_ATCC_1997 TTFPGDIFALIVGVSLLFAHLERSGTLSWFVELVYGAVGDRHVLLPWAGF
MSMEG_4611|M.smegmatis_MC2_155 AGFPAGLFVILVGVTYLFAIAKANGAIDWLVDTAVTLVRGRVWAFPWIMF
: **..:*.::***: *** : .*::.*:*: . * .* :** *
MAB_4247|M.abscessus_ATCC_1997 LVGAILSTAGAFSTAVIAFLVPLVAHVAVRYRSSFYLCELGVVIGANSAG
MSMEG_4611|M.smegmatis_MC2_155 VLAAGLAGVGAVPPAAVAIIAPTAMRIAHRYGISPLLMGLMVANGVSAGE
::.* *: .**...*.:*::.* . ::* ** * * * *. *..:.
MAB_4247|M.abscessus_ATCC_1997 LSPLNPTGAVVRAAATKFHVDYPGWALWAVSMVVAALVVLCLQLLDAFRR
MSMEG_4611|M.smegmatis_MC2_155 FSPIGLFGIIVNDVAVSNGVTSSPILLFASCFAVNAIMCYVLCAVINRRA
:**:. * :*. .*.. * . *:* .:.* *:: * : *
MAB_4247|M.abscessus_ATCC_1997 RRGGGFDLTPAPAAAT-------------ETDAAESPSLRYAWCAGLA-L
MSMEG_4611|M.smegmatis_MC2_155 RRHDKTDDSIGTGGPTPPRGGSSVRVVDADVAVAVRPPMTRSIAATLAGL
** . * : .....* :. .* *.: : .* ** *
MAB_4247|M.abscessus_ATCC_1997 LTFVLLVVVVKTDVGVTAIALAALMQIAFHPHERSLLARIPWPSILLLCG
MSMEG_4611|M.smegmatis_MC2_155 LVMVGIVACTELDIGFVAMTLAVILSVLSPDAAKEALPKIAWPSVFLIVG
*.:* :*. .: *:*..*::**.::.: :. *.:*.***::*: *
MAB_4247|M.abscessus_ATCC_1997 LLTYLGLMQKIGTIDSIASVLHRLGTGAVLILVLAYLTALLCNIESSTLG
MSMEG_4611|M.smegmatis_MC2_155 IVTYVGLLQELGTVTYVSDAVAHLQS-PLLIALLICVIGAVVSAFASTTG
::**:**:*::**: ::..: :* : .:** :* : . : . :** *
MAB_4247|M.abscessus_ATCC_1997 VLGLMMPIVFS-TFTEPAQMFWVVCAVCVPAALMVMNPIHVAGTLIIGNS
MSMEG_4611|M.smegmatis_MC2_155 ILGALVPLAVPFLLSGEVSSIAMIIALSLSSSIVDASPFSTSGALIVANA
:** ::*:... :: .. : :: *:.:.:::: .*: .:*:**:.*:
MAB_4247|M.abscessus_ATCC_1997 AADEQDNLFRRLLALAGSLALVVPGLLSIVPIALA-
MSMEG_4611|M.smegmatis_MC2_155 AEEDRDRVMSHLLRWGMSMIVVAPVLSWAVFVLPGW
* :::*.:: :** . *: :*.* * * : .