For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTLPKQPFGRDASLDGQEVELDFAREWVEFYDPENPEHLIKADMTWLLSHWTCVFGTPACQGTVAGRPD DGCCSHGAFLSDADDRANLDDSVKKLTDADWQFREKGLGRKGYLEKDELDGEKQWRTRKYKGACIFLNRP GFAGGIGCALHSKALKLGVEPLTMKPEVCWQLPIRRTQAWVDRPDGSQILKTTITEYDRRGWGEGGADLH WYCTGDPAAHVGKKQVWQSYAPELTELLGEKAYAELAAMCKRRKGLGLIAVHPATVAAAPH
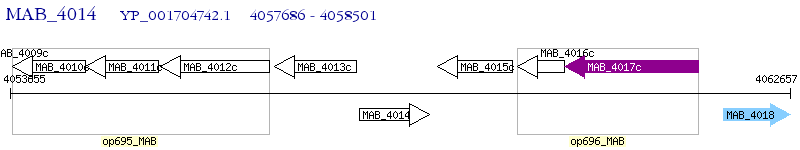
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4014 | - | - | 100% (271) | hypothetical protein MAB_4014 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0506c | - | 1e-131 | 84.92% (252) | hypothetical protein Mb0506c |
| M. gilvum PYR-GCK | Mflv_0080 | - | 1e-132 | 84.80% (250) | hypothetical protein Mflv_0080 |
| M. tuberculosis H37Rv | Rv0495c | - | 1e-131 | 84.92% (252) | hypothetical protein Rv0495c |
| M. leprae Br4923 | MLBr_02435 | - | 1e-122 | 77.78% (252) | hypothetical protein MLBr_02435 |
| M. marinum M | MMAR_0821 | - | 1e-132 | 84.52% (252) | hypothetical protein MMAR_0821 |
| M. avium 104 | MAV_4657 | - | 1e-132 | 85.54% (249) | hypothetical protein MAV_4657 |
| M. smegmatis MC2 155 | MSMEG_0938 | - | 1e-130 | 84.62% (247) | hypothetical protein MSMEG_0938 |
| M. thermoresistible (build 8) | TH_0880 | - | 1e-130 | 85.02% (247) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4565 | - | 1e-132 | 84.13% (252) | hypothetical protein MUL_4565 |
| M. vanbaalenii PYR-1 | Mvan_0832 | - | 1e-132 | 85.43% (247) | hypothetical protein Mvan_0832 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0821|M.marinum_M --------------------------------MANSCLKTVQPGQEVELD
MUL_4565|M.ulcerans_Agy99 --------------------------------MANSCLKTVQPGQEIELD
Mb0506c|M.bovis_AF2122/97 MWRPAQGARWHVPAVLGYGGIPRRASWSNVESVANSRRRPVHPGQEVELD
Rv0495c|M.tuberculosis_H37Rv MWRPAQGARWHVPAVLGYGGIPRRASWSNVESVANSRRRPVHPGQEVELD
MLBr_02435|M.leprae_Br4923 -------------------MSSCPESGSTFEYVANSHLEPVHPGEEVDLD
MAV_4657|M.avium_104 -----------------------------MDDVANSHPQPGQPGQEVELD
Mflv_0080|M.gilvum_PYR-GCK --------------------------------MKTVSASASHPG-EVELD
Mvan_0832|M.vanbaalenii_PYR-1 --------------------------------MKTVSASASRPG-EVELD
MSMEG_0938|M.smegmatis_MC2_155 -------------------------------------MTRVHPG-EVELD
TH_0880|M.thermoresistible__bu ------------------------------------------VG-EVELD
MAB_4014|M.abscessus_ATCC_1997 ---------------------------MTTLPKQPFGRDASLDGQEVELD
* *::**
MMAR_0821|M.marinum_M FPREWVEFYDPDNPEHLIAADLTWLLSHWSCVFGTPACQGTVAGRPHDGC
MUL_4565|M.ulcerans_Agy99 FPREWVEFYDPDNPEHLIAADLTWLLSHWSCVFGTPACQGTVAGRPNDGC
Mb0506c|M.bovis_AF2122/97 FAREWVEFYDPDNPEHLIAADLTWLLSRWACVFGTPACQGTVAGRPNDGC
Rv0495c|M.tuberculosis_H37Rv FAREWVEFYDPDNPEHLIAADLTWLLSRWACVFGTPACQGTVAGRPNDGC
MLBr_02435|M.leprae_Br4923 FTREWVEFYDPDNSEQLIAADLTWLLSRWTCVFGTPACRGTVAGRPDDGC
MAV_4657|M.avium_104 FAREWVEFYDPDNPEHLIAADLTWLLSRWTCVFGTPACRGTVAGRPDDGC
Mflv_0080|M.gilvum_PYR-GCK FAREWVEFYDPDNPEHLIAADMTWLLSRWTCVFGTPACKGTVVDRPDDGC
Mvan_0832|M.vanbaalenii_PYR-1 FAREWVEFYDPDNPEHLIAADMTWLLSRWTCVFGTPACKGTVEDRPDDGC
MSMEG_0938|M.smegmatis_MC2_155 FAREWVEFYDPDDADHLIAADMTWLLSRWTCVFGTPACQGTVEGRPDDGC
TH_0880|M.thermoresistible__bu FAREWVEFYDPDNPEHLIAADMTWLLSRWTCVFGTPACQGTVARRPDDGC
MAB_4014|M.abscessus_ATCC_1997 FAREWVEFYDPENPEHLIKADMTWLLSHWTCVFGTPACQGTVAGRPDDGC
*.*********::.::** **:*****:*:********:*** **.***
MMAR_0821|M.marinum_M CSHGAFLSDDDDRARLDDAVKKLTDDDWQYRDKGLGRKGYLELDEHDDQP
MUL_4565|M.ulcerans_Agy99 CSHGAFLSDDDDRARLDDAVKKLTDDDWQYRDKGLGRKGYLELDEHDDQP
Mb0506c|M.bovis_AF2122/97 CSHGAFLSDDDDRTRLADAVHKLTDDDWQFRAKGLRRKGYLELDEHDGQP
Rv0495c|M.tuberculosis_H37Rv CSHGAFLSDDDDRTRLADAVHKLTDDDWQFRAKGLRRKGYLELDEHDGQP
MLBr_02435|M.leprae_Br4923 CSHGAFLSDDADRTRLDDAVKKLSHDDWQFREKGLGRKGYLELDEHDGQS
MAV_4657|M.avium_104 CSHGAFLSDDDDRARLDDAVKQLTDDDWQFREKGLGRKGYLELDEHDGQP
Mflv_0080|M.gilvum_PYR-GCK CSHGAFLSDDDDRAQLDDAVTLLTAEDWQYRDKGLGKKGYLEMDEYDGKP
Mvan_0832|M.vanbaalenii_PYR-1 CSHGAFLSDDDDRAQLDDAVKLLTDEDWQFRDKGLGRKGYLELDEYDDKP
MSMEG_0938|M.smegmatis_MC2_155 CSHGAFLSDDDDRARLDDAVKQLTDEDWQFREKGLGKKGYLEDDEYDGKP
TH_0880|M.thermoresistible__bu CSHGAFLTDDDDRARLDAAVDQLTDEDWQFRDKGLGKKGYLEEDEYDGEA
MAB_4014|M.abscessus_ATCC_1997 CSHGAFLSDADDRANLDDSVKKLTDADWQFREKGLGRKGYLEKDELDGEK
*******:* **:.* :* *: ***:* *** :***** ** *.:
MMAR_0821|M.marinum_M QYRTRKHKDACIFLNRPGFAGGTGCALHSKALKLGVAPLTMKPDVCWQLP
MUL_4565|M.ulcerans_Agy99 QYRTRKHKDACIFLNRPGFAGGTGCALHSKALKLGVAPLTMKPDVCWQLP
Mb0506c|M.bovis_AF2122/97 QHRTRKHKGACIFLNRPGFAGGAGCALHSKALKLGVPPLTMKPDVCWQLP
Rv0495c|M.tuberculosis_H37Rv QHRTRKHKGACIFLNRPGFAGGAGCALHSKALKLGVPPLTMKPDVCWQLP
MLBr_02435|M.leprae_Br4923 QFRTRKHKNACIFLNRPGFPIGAGCALHSKALKLGVPPRTMKPDICWQLP
MAV_4657|M.avium_104 QYRTRKHKDACIFLNRPGFAGGVGCALHSKALKLGVPPLTMKPDVCWQLP
Mflv_0080|M.gilvum_PYR-GCK NWRTRKYKGACIFLNRPGWPKGIGCALHSKALEIGVEPLTLKPEVCWQLP
Mvan_0832|M.vanbaalenii_PYR-1 NWRTRKYKGACIFLNRPGWPKGIGCALHQKALAIGVEPLTLKPEVCWQLP
MSMEG_0938|M.smegmatis_MC2_155 NLRTRKYKGACIFLNRPGFPTGIGCALHSKALKLGVEPWTMKPDVCWQLP
TH_0880|M.thermoresistible__bu ALRTRKYKGACIFLNRPGFPGGIGCALHIKALKLGVEPLTMKPDVCWQLP
MAB_4014|M.abscessus_ATCC_1997 QWRTRKYKGACIFLNRPGFAGGIGCALHSKALKLGVEPLTMKPEVCWQLP
****:*.*********:. * ***** *** :** * *:**::*****
MMAR_0821|M.marinum_M IRRSQEWVTRPDGSEILKTTLTEYDRRGWGSGGADLHWYCTGDPAAHVGS
MUL_4565|M.ulcerans_Agy99 IRRSQEWVTRPDGSEILKTTLTEYDRRGWGSGGADLHWYCTGDPAAHVGS
Mb0506c|M.bovis_AF2122/97 IRRSQEWVTRPDGTEILKTTLTEYDRRGWGSGGADLHWYCTGDPAAHVGT
Rv0495c|M.tuberculosis_H37Rv IRRSQEWVTRPDGTEILKTTLTEYDRRGWGSGGADLHWYCTGDPAAHVGT
MLBr_02435|M.leprae_Br4923 IRHSQEWVTRPDGTEILKTTVTEYDRRSWGSGGADLHWYCTGDPASHVDS
MAV_4657|M.avium_104 IRRSQEWVTRPDGTEILKTWVTEYDRRGWGSGGADLHWYCTGDPAAHVGA
Mflv_0080|M.gilvum_PYR-GCK IRRTQEWITRPDDTEILKTTITEYDRRGWGEGGADLHWYCTGDPAAHVGA
Mvan_0832|M.vanbaalenii_PYR-1 IRRTQEWITRPDGSEILKTTITEYDRRGWGEGGLDLHWYCTGDPNTHVGA
MSMEG_0938|M.smegmatis_MC2_155 IRRSQDWVERPDGTQILKTVITEYDRRGWGEGGADLHWYCTGDPNAHVGS
TH_0880|M.thermoresistible__bu IRRSQEWVTRPDGTEVLKTTITEYDRRGWGEGGHDLHWYCTGDPAAHVGT
MAB_4014|M.abscessus_ATCC_1997 IRRTQAWVDRPDGSQILKTTITEYDRRGWGEGGADLHWYCTGDPAAHVGK
**::* *: ***.:::*** :******.**.** ********** :**.
MMAR_0821|M.marinum_M KQVWESLADELTELLGAKAYAELAAVCKRRSQLGLVAVHPATRAAE----
MUL_4565|M.ulcerans_Agy99 KQVWESLADELTELLGAKAYAELAAVCKRRSQLGLVAVHPATRAAE----
Mb0506c|M.bovis_AF2122/97 KQVWQSLADELTELLGEKAYGELAAMCKRRSQLGLIAVHPATRAAQ----
Rv0495c|M.tuberculosis_H37Rv KQVWQSLADELTELLGEKAYGELAAMCKRRSQLGLIAVHPATRAAQ----
MLBr_02435|M.leprae_Br4923 KQLWESLADELTELLGAKAYAKLAAICKRRNRLGIIAVHPATQEAK----
MAV_4657|M.avium_104 TQVWESLADELTELLGAKAYAELAAMCKRRSKLGLIAVHPATRIAE----
Mflv_0080|M.gilvum_PYR-GCK KPVWQSYAPELTELLGEKAYAELAAMCKRRSGLGLIAIHPATRAAE----
Mvan_0832|M.vanbaalenii_PYR-1 KQVWESYAPELTELLGEKTYAELAAMCKRRSGLGLIAVHPATRLAE----
MSMEG_0938|M.smegmatis_MC2_155 RPVWESYAPELTALLGEKAYAELAAICKRRSRLGLVAVHPATRLAESGTS
TH_0880|M.thermoresistible__bu RPVWQSYAAELTELLGEKAYAELAAICKRRGKLGLVAVHPATRLAE----
MAB_4014|M.abscessus_ATCC_1997 KQVWQSYAPELTELLGEKAYAELAAMCKRRKGLGLIAVHPATVAAAPH--
:*:* * *** *** *:*.:***:**** **::*:**** *
MMAR_0821|M.marinum_M ----
MUL_4565|M.ulcerans_Agy99 ----
Mb0506c|M.bovis_AF2122/97 ----
Rv0495c|M.tuberculosis_H37Rv ----
MLBr_02435|M.leprae_Br4923 ----
MAV_4657|M.avium_104 ----
Mflv_0080|M.gilvum_PYR-GCK ----
Mvan_0832|M.vanbaalenii_PYR-1 ----
MSMEG_0938|M.smegmatis_MC2_155 GPQR
TH_0880|M.thermoresistible__bu ----
MAB_4014|M.abscessus_ATCC_1997 ----