For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MYYLALLIGPQQERTPDEAAQELTDYLNFQAKAASSIRAGDALYPEADSVRVTGGPDAPVITDGPFAEGA EVANGYYVLEADNLDDALALARDIPEAKRGAVEVWPLVDCSREFGGKGSTSWLALLLEPADRAYVPGSPE WEAVAARHGVFGEKASGHIKIAGPLHPPSTATTIRVRDGEAVLTDGPYIEGTEIANGFYVITTDDRDSAI RLASQIPASAVQLRQLTGVSGF
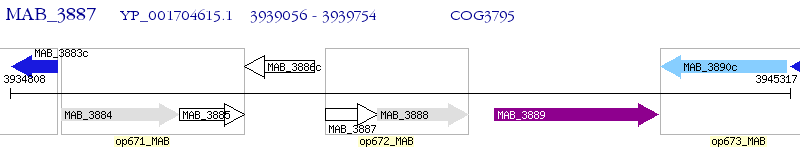
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3887 | - | - | 100% (232) | hypothetical protein MAB_3887 |
| M. abscessus ATCC 19977 | MAB_4454c | - | 7e-08 | 32.35% (102) | hypothetical protein MAB_4454c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0976 | - | 2e-79 | 60.61% (231) | hypothetical protein MMAR_0976 |
| M. avium 104 | MAV_4518 | - | 9e-74 | 58.70% (230) | DGPF domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_1349 | - | 5e-73 | 57.83% (230) | DGPF domain-containing protein |
| M. thermoresistible (build 8) | TH_0191 | - | 7e-06 | 27.45% (102) | dgpf protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3354 | - | 9e-69 | 53.25% (231) | DGPFAETKE family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_4518|M.avium_104 ----------------------------MQYFALLISREQER-------K
MSMEG_1349|M.smegmatis_MC2_155 ----------------------------MLYFALMKSNESQR-------T
MMAR_0976|M.marinum_M ----------------------------MHYFALLISKERDR-------T
Mvan_3354|M.vanbaalenii_PYR-1 MSDSCGAVRRSGREWNTRVPLQAPEDITMYYFALLQVPERDL-------T
MAB_3887|M.abscessus_ATCC_1997 ----------------------------MYYLALLIGPQQER-------T
TH_0191|M.thermoresistible__bu ----------------------------MARYMLIMRSTPEAGQEMENVD
* *: :
MAV_4518|M.avium_104 PDDAAAAMAAWESFHAKAGPAIKAGDALAPAAAAAVITGGPDAPVVTDGP
MSMEG_1349|M.smegmatis_MC2_155 PEKAAEGFAAFQAFHAKAASALLAGDALAPGSRAVRIEGGPDAPVVTDGP
MMAR_0976|M.marinum_M PSEGAAEMAAYEAFHAQAASAIRGGDALLSAADAAQITGGPNAPVITDGP
Mvan_3354|M.vanbaalenii_PYR-1 PEQRESEMRAYTDFHALAARAILAGDALASATEAVRISGGPDNPMVTDGP
MAB_3887|M.abscessus_ATCC_1997 PDEAAQELTDYLNFQAKAASSIRAGDALYPEADSVRVTGGPDAPVITDGP
TH_0191|M.thermoresistible__bu FDEIIAAMGRYNEELIKAG-VMRGGEGLAGPEEGFVVNFDQNPPVVTDGP
.. : : *. : .*:.* . : . : *::****
MAV_4518|M.avium_104 FAETAEVACGYYIFEAENLDEALALARDVPVAAFGAVELWPVVHAVEPSR
MSMEG_1349|M.smegmatis_MC2_155 FAEASEIACGYYVLEADNLDDALALAREIPVAAFGAVEIWPIVHFQAPTQ
MMAR_0976|M.marinum_M FAEGAEVAGGYYVFETENLDEVLELARAIPAAKYGAVEVWPMAGPGLPTQ
Mvan_3354|M.vanbaalenii_PYR-1 FAEGAEVAGGFYVFEADNLDDALKLAEQIPAAGYGAVEVWPMVHWNAPDR
MAB_3887|M.abscessus_ATCC_1997 FAEGAEVANGYYVLEADNLDDALALARDIPEAKRGAVEVWPLVDCSREFG
TH_0191|M.thermoresistible__bu YPDARELFNGFWILEVSSKDEARQWAAKCPLGPGARLEVRRIN-------
:.: *: *::::*... *:. * * . . :*: :
MAV_4518|M.avium_104 RITGNDWLALLLEPAESAHTPGTPEWEAVAAKHAELHAAAGDHIIGGAAL
MSMEG_1349|M.smegmatis_MC2_155 PIG-NDWLALLLEPPENTRTPGTDEWNELAGRHVELSSTIGDRTLAGAPL
MMAR_0976|M.marinum_M PLQGSNWIALLLEPPERAVAPGTPEWEAMAAQHQQFGAAASEHIKAGAGL
Mvan_3354|M.vanbaalenii_PYR-1 PTTGNDWLALLLEPAEQVTTPCTPEWDEGVCRHADFNAAAGEHVLGGAPL
MAB_3887|M.abscessus_ATCC_1997 GKGSTSWLALLLEPADRAYVPGSPEWEAVAARHGVFGEKASGHIKIAGPL
TH_0191|M.thermoresistible__bu ---------------DVEDFPQDNEWIQKEIRYREEGVWA----------
: * ** ::
MAV_4518|M.avium_104 HDKSTATTVRVRDGEVLITDGPYVESAEIATGIYLLGAADRDEAVKIASM
MSMEG_1349|M.smegmatis_MC2_155 HPPSTATTVRVRDGEVMITDGPYIEGAEVANGFYVLRAPDRDEAVKLASM
MMAR_0976|M.marinum_M HPPSTATTVRVRDGQVLITDGPYVEGAEVANGYYVLSAADRDEAVKLASM
Mvan_3354|M.vanbaalenii_PYR-1 HPPTTATTVRVRGTEVLMTDGPFAESAEVANGFYLLRAADRDEAAKVAAM
MAB_3887|M.abscessus_ATCC_1997 HPPSTATTIRVRDGEAVLTDGPYIEGTEIANGFYVITTDDRDSAIRLASQ
TH_0191|M.thermoresistible__bu --------------------------------------------------
MAV_4518|M.avium_104 IPASTVQLRQLAGVSDL
MSMEG_1349|M.smegmatis_MC2_155 LPASAVELRQLAGVSNL
MMAR_0976|M.marinum_M IPASTVLVRGLAGVSGL
Mvan_3354|M.vanbaalenii_PYR-1 IPAGAVQVRRLAGVAGL
MAB_3887|M.abscessus_ATCC_1997 IPASAVQLRQLTGVSGF
TH_0191|M.thermoresistible__bu -----------------