For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TQVFDDNNRVVPVTVVKAGPNVVTRIRTTEQDGYSAVQLAYGEISPRKVTKPVTGQFAAAGINPRRHLAE LRLDDEAAAAEYEVGQELTAEIFSDGAYVDVTGTSKGKGFAGTMKRHGFRGQGASHGAQAVHRRPGSIGG CATPGRVFKGTRMSGRMGSDRVTTQNLVVHKVDAENGVLLIKGAIPGRKGGLVVVRTAVKRGEK*
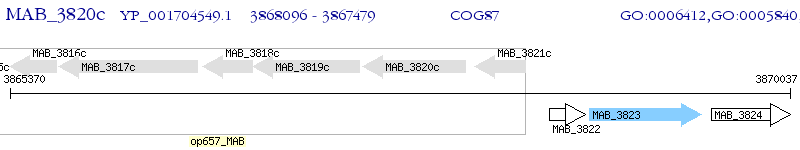
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3820c | rplC | - | 100% (205) | 50S ribosomal protein L3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0721 | rplC | 1e-103 | 86.83% (205) | 50S ribosomal protein L3 |
| M. gilvum PYR-GCK | Mflv_5049 | rplC | 1e-106 | 92.23% (206) | 50S ribosomal protein L3 |
| M. tuberculosis H37Rv | Rv0701 | rplC | 1e-103 | 86.83% (205) | 50S ribosomal protein L3 |
| M. leprae Br4923 | MLBr_01863 | rplC | 1e-103 | 86.83% (205) | 50S ribosomal protein L3 |
| M. marinum M | MMAR_1031 | rplC | 1e-103 | 87.32% (205) | 50S ribosomal protein L3 RplC |
| M. avium 104 | MAV_4471 | rplC | 1e-105 | 89.76% (205) | 50S ribosomal protein L3 |
| M. smegmatis MC2 155 | MSMEG_1436 | rplC | 1e-109 | 93.17% (205) | 50S ribosomal protein L3 |
| M. thermoresistible (build 8) | TH_0407 | rplC | 1e-103 | 86.83% (205) | PROBABLE 50S RIBOSOMAL PROTEIN L3 RPLC |
| M. ulcerans Agy99 | MUL_0790 | rplC | 1e-103 | 86.83% (205) | 50S ribosomal protein L3 |
| M. vanbaalenii PYR-1 | Mvan_1305 | rplC | 1e-110 | 94.15% (205) | 50S ribosomal protein L3 |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3820c|M.abscessus_ATCC_199 -------------MTQVFDDNNRVVPVTVVKAGPNVVTRIRTTEQDGYSA
MSMEG_1436|M.smegmatis_MC2_155 -MARKGILGTKLGMTQVFDENNKVVPVTVVKAGPNVVTRIRTTERDGYSA
Mflv_5049|M.gilvum_PYR-GCK -MARKGILGTKLGMTQVFDENNKVVPVTVVKAGPNVVTRIRTTERDGYSA
Mvan_1305|M.vanbaalenii_PYR-1 -MARKGILGTKLGMTQVFDENNKVVPVTVVKAGPNVVTRIRTPERDGYSA
TH_0407|M.thermoresistible__bu -MARKGILGTKLGMTQVFDENNKVVPVTVVKAGPNVVTRIRTPERDGYSA
Mb0721|M.bovis_AF2122/97 -MARKGILGTKLGMTQVFDESNRVVPVTVVKAGPNVVTRIRTPERDGYSA
Rv0701|M.tuberculosis_H37Rv -MARKGILGTKLGMTQVFDESNRVVPVTVVKAGPNVVTRIRTPERDGYSA
MLBr_01863|M.leprae_Br4923 MMARKGILGTKLGMTQVFDENNRVVPVTVVKAGPNVVTRIRTLERDGYSA
MAV_4471|M.avium_104 -MARKGILGTKLGMTQVFDENNRVVPVTVVKAGPNVVTRIRTPEQDGYSA
MMAR_1031|M.marinum_M -MARKGILGTKLGMTQVFDENNKVVPVTVVKAGPNVVTRIRTPERDGYSA
MUL_0790|M.ulcerans_Agy99 -MARKGILGTKLGMTQVFDENNKVVPVTVVKAGPNVVTRIRTPERDGYSA
******:.*:******************* *:*****
MAB_3820c|M.abscessus_ATCC_199 VQLAYGEISPRKVTKPVTGQFAAAGINPRRHLAELRLDD-EAAAAEYEVG
MSMEG_1436|M.smegmatis_MC2_155 VQLAYGEISPRKVIKPVAGQFAAAGVNPRRHVAELRLDD-EAAVAEYEVG
Mflv_5049|M.gilvum_PYR-GCK VQLAYGEISPRKVNKPVTGQFAAAGVNPRRHLAELRLADNEDAGALYEVG
Mvan_1305|M.vanbaalenii_PYR-1 VQLAYGEISPRKVNKPVTGQYAAAGVNPRRHLAELRLDD-EAAAAEYEVG
TH_0407|M.thermoresistible__bu VQLAYGEINPRKVNKPLAGQFAAAGVNPRRHIAELRLDD-EAAATEYEVG
Mb0721|M.bovis_AF2122/97 VQLAYGEISPRKVNKPLTGQYTAAGVNPRRYLAELRLDD-SDAATEYQVG
Rv0701|M.tuberculosis_H37Rv VQLAYGEISPRKVNKPLTGQYTAAGVNPRRYLAELRLDD-SDAATEYQVG
MLBr_01863|M.leprae_Br4923 VQLAYGEISPRKVNKPVTGQYNSAGVNPRRYLAELRLDH-PDAAAEYEVG
MAV_4471|M.avium_104 VQLAYGEISPRKVNKPVTGQYAAAGINPRRFLAELRLDN-PDAAAEYQVG
MMAR_1031|M.marinum_M VQLAYGEISPRKVNKPVTGQYTAAGVNPRRHLAELRLDD-AEAVTEYEVG
MUL_0790|M.ulcerans_Agy99 VQLAYGEISPRKVNKPVTGQYTAAGVNPRRHLAELRLDD-AEAVTEYEVG
********.**** **::**: :**:****.:***** . * : *:**
MAB_3820c|M.abscessus_ATCC_199 QELTAEIFSDGAYVDVTGTSKGKGFAGTMKRHGFRGQGASHGAQAVHRRP
MSMEG_1436|M.smegmatis_MC2_155 QELTAEIFSDGAYVDVTGTSKGKGFAGTMKRHGFRGQGAAHGAQAVHRRP
Mflv_5049|M.gilvum_PYR-GCK QELTAEIFADGAYVDVTGTSKGKGFAGTMKRHGFKGQGASHGAQAVHRRP
Mvan_1305|M.vanbaalenii_PYR-1 QELTAEIFADGAYVDVTGTSKGKGFAGTMKRHGFRGQGASHGAQAVHRRP
TH_0407|M.thermoresistible__bu QELTADIFTEGSYVDVTGTSKGKGFAGTMKRHGFAGQGASHGTQAVHRRP
Mb0721|M.bovis_AF2122/97 QELTAEIFADGSYVDVTGTSKGKGFAGTMKRHGFRGQGASHGAQAVHRRP
Rv0701|M.tuberculosis_H37Rv QELTAEIFADGSYVDVTGTSKGKGFAGTMKRHGFRGQGASHGAQAVHRRP
MLBr_01863|M.leprae_Br4923 QELTAEIFADATYVDVTGTSKGKGFSGTMKRHGFRGQGASHGAQAVHRRP
MAV_4471|M.avium_104 QELTAEIFTDGSYVDVTGTSKGKGFAGTMKRHGFRGQGASHGAQAVHRRP
MMAR_1031|M.marinum_M QELTAEIFADGSYVDVTGTSKGKGFAGTMKRHGFSGQGASHGAQAVHRRP
MUL_0790|M.ulcerans_Agy99 QELTAEIFADGSYVDVTGTSKGKGFAGTMKRHGFSGQGASHGAQAVHRRP
*****:**::.:*************:******** ****:**:*******
MAB_3820c|M.abscessus_ATCC_199 GSIGGCATPGRVFKGTRMSGRMGSDRVTTQNLVVHKVDAENGVLLIKGAI
MSMEG_1436|M.smegmatis_MC2_155 GSIGGCATPGRVFKGTRMSGRMGNDRVTTQNLKVHKVDAENGVLLIKGAI
Mflv_5049|M.gilvum_PYR-GCK GSIGGCATPGRVFKGTRMSGRMGSDRVTTQNLKVHKVDAENGVLLIKGAI
Mvan_1305|M.vanbaalenii_PYR-1 GSIGGCATPGRVFKGTRMSGRMGSDRVTTQNLKVHKVDAENGVLLIKGAI
TH_0407|M.thermoresistible__bu GSIGGCATPGRVFKGTRMAGRMGNERVTTLNLKVHKVDAENGVLLIKGAI
Mb0721|M.bovis_AF2122/97 GSIGGCATPARVFKGTRMAGRMGNDRVTVLNLLVHKVDAENGVLLIKGAV
Rv0701|M.tuberculosis_H37Rv GSIGGCATPARVFKGTRMAGRMGNDRVTVLNLLVHKVDAENGVLLIKGAV
MLBr_01863|M.leprae_Br4923 GSIGGCATPARVFKGTRMAGRMGNDRVTVQNLLVHKVDTENGVLLIKGAV
MAV_4471|M.avium_104 GSIGGCATPARVFKGTRMAGRMGNDRVTVQNLLVHKVDTENGVLLIKGAV
MMAR_1031|M.marinum_M GSIGGCATPARVFKGTRMAGRMGNDRVTVQNLLVHKVDAEQGVLLIKGAV
MUL_0790|M.ulcerans_Agy99 GSIGGCATPARVFKGTRMAGRMGNDRVTVQNLLVHKVDAEQSVLLIKGAV
*********.********:****.:***. ** *****:*:.*******:
MAB_3820c|M.abscessus_ATCC_199 PGRKGGLVVVRTAVKRGEK-
MSMEG_1436|M.smegmatis_MC2_155 PGRNGGLVVVRSAIKRGEK-
Mflv_5049|M.gilvum_PYR-GCK PGRNGGLVVVRSAIKRGEK-
Mvan_1305|M.vanbaalenii_PYR-1 PGRNGGLVVVRSAIKRGEKA
TH_0407|M.thermoresistible__bu PGRNGGLVMVRSAIKRGDKK
Mb0721|M.bovis_AF2122/97 PGRTGGLVMVRSAIKRGEK-
Rv0701|M.tuberculosis_H37Rv PGRTGGLVMVRSAIKRGEK-
MLBr_01863|M.leprae_Br4923 PGRTGGLVMVRSAIKRGEK-
MAV_4471|M.avium_104 PGRTGGLVMVRSAVKRGEK-
MMAR_1031|M.marinum_M PGRTGGLVMVRSAIKRGEK-
MUL_0790|M.ulcerans_Agy99 PGRTGGLVTVRSAIKRGEK-
***.**** **:*:***:*