For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRIGKQPVPVPAGVDVNIDGQNISVKGSKGTLELTVSEPISVSRNDDGAIVVTRPDDERRNRSLHGLSRT LIANLVTGVTQGYTTKMEIFGVGYRVVAKGSNLEFALGYSHPVLIEAPEGITFAVETPTKFSISGIDKQK VGQISANIRRLRRPDPYKGKGIRYEGEQIRRKVGKTGK*
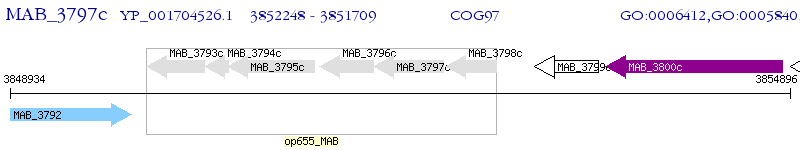
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3797c | rplF | - | 100% (179) | 50S ribosomal protein L6 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0740 | rplF | 6e-91 | 88.83% (179) | 50S ribosomal protein L6 |
| M. gilvum PYR-GCK | Mflv_5032 | rplF | 6e-91 | 91.06% (179) | 50S ribosomal protein L6 |
| M. tuberculosis H37Rv | Rv0719 | rplF | 6e-91 | 88.83% (179) | 50S ribosomal protein L6 |
| M. leprae Br4923 | MLBr_01844 | rplF | 6e-89 | 85.47% (179) | 50S ribosomal protein L6 |
| M. marinum M | MMAR_1050 | rplF | 6e-88 | 85.47% (179) | 50S ribosomal protein L6, RplF |
| M. avium 104 | MAV_4450 | rplF | 2e-92 | 91.06% (179) | 50S ribosomal protein L6 |
| M. smegmatis MC2 155 | MSMEG_1470 | rplF | 2e-88 | 88.83% (179) | 50S ribosomal protein L6 |
| M. thermoresistible (build 8) | TH_0308 | rplF | 1e-86 | 85.00% (180) | PROBABLE 50S RIBOSOMAL PROTEIN L6 RPLF |
| M. ulcerans Agy99 | MUL_0809 | rplF | 4e-88 | 85.47% (179) | 50S ribosomal protein L6 |
| M. vanbaalenii PYR-1 | Mvan_1337 | rplF | 4e-90 | 89.94% (179) | 50S ribosomal protein L6 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1050|M.marinum_M MSRIGKQPVPVPTGVDVMIEGQKVSVKGPKGALDLTVAEPITVSRNDDGA
MUL_0809|M.ulcerans_Agy99 MSRIGKQPVPVPTGVDVMIEGQKVSVKGPKGALDLTVAEPITVSRNDDGA
MAV_4450|M.avium_104 MSRIGKQPVPVPAGVDVTIDGQKVSVKGPKGTLDLTVAEPITVSRNDDGA
Mb0740|M.bovis_AF2122/97 MSRIGKQPIPVPAGVDVTIEGQSISVKGPKGTLGLTVAEPIKVARNDDGA
Rv0719|M.tuberculosis_H37Rv MSRIGKQPIPVPAGVDVTIEGQSISVKGPKGTLGLTVAEPIKVARNDDGA
MLBr_01844|M.leprae_Br4923 MSRIGKQPIPVPAGVDITIDGQNVLVKGPKGTLDLTVAEPIMLARNDEGA
TH_0308|M.thermoresistible__bu MSRIGKQPIPVPAGVDVTIDGQRVSVKGPKGTLSLDVVEPIKVSRNDDGA
Mflv_5032|M.gilvum_PYR-GCK MSRIGKQPVPVPAGVDVTISGQNVSVKGPKGTLTLDVAEPIEVSRNDDGA
Mvan_1337|M.vanbaalenii_PYR-1 MSRIGKQPVTVPSGVDVTIDGQNVSVKGPKGTLTLDVAEPIAVTRDDDGA
MAB_3797c|M.abscessus_ATCC_199 MSRIGKQPVPVPAGVDVNIDGQNISVKGSKGTLELTVSEPISVSRNDDGA
MSMEG_1470|M.smegmatis_MC2_155 MSRIGKQPVPVPSGVDVTINGQNLSVKGPKGTLTLDVAEPISVSRAEDGA
********:.**:***: *.** : ***.**:* * * *** ::* ::**
MMAR_1050|M.marinum_M IVISRPNDERRNRSLHGLSRTLVSNLVTGVTEGYTTKMEIHGVGYRVQLK
MUL_0809|M.ulcerans_Agy99 IVISRPNDERRNRSLHGLSRTLVSNLVTGVTEGYTTKMEIHGVGYRVQLK
MAV_4450|M.avium_104 IVVTRPNDERRNRSLHGLSRTLVSNLVTGVTQGYTTKMEIFGVGYRVQLK
Mb0740|M.bovis_AF2122/97 IVVTRPDDERRNRSLHGLSRTLVSNLVTGVTQGYTTKMEIFGVGYRVQLK
Rv0719|M.tuberculosis_H37Rv IVVTRPDDERRNRSLHGLSRTLVSNLVTGVTQGYTTKMEIFGVGYRVQLK
MLBr_01844|M.leprae_Br4923 IVVTRPDNERRNRSLHGLSRTLVSNLVTGVTQGYTVSMEIFGVGYRAQLK
TH_0308|M.thermoresistible__bu IVVTRPDDERRNRSLHGLSRSLVANLITGVTEGYTTRMEIHGVGYRVQAK
Mflv_5032|M.gilvum_PYR-GCK IVVTRPNDERRNRSLHGLSRTLIANLVTGVTEGYTTKMEIFGVGYRVVAK
Mvan_1337|M.vanbaalenii_PYR-1 IVVTRPNDERRNRSLHGLSRTLIANLVTGVTEGYTTKMEIFGVGYRVVAK
MAB_3797c|M.abscessus_ATCC_199 IVVTRPDDERRNRSLHGLSRTLIANLVTGVTQGYTTKMEIFGVGYRVVAK
MSMEG_1470|M.smegmatis_MC2_155 IVVTRPDDERRSRSLHGLSRTLIANLVTGVTEGYTQKMEIFGVGYRVQLK
**::**::***.********:*::**:****:*** ***.*****. *
MMAR_1050|M.marinum_M GAN-LEFALGYSHPVVIEAPEGITFAVQAPTKFTVSGIDKQKVGQISANI
MUL_0809|M.ulcerans_Agy99 GAN-LEFALGYSHPVVIEAPEGITFAVQAPTKFTVSGIDKQKVGQISANI
MAV_4450|M.avium_104 GSN-LEFALGYSHPVVIEAPEGITFAVQSPTKFSISGIDKQKVGQISANI
Mb0740|M.bovis_AF2122/97 GSN-LEFALGYSHPVVIEAPEGITFAVQAPTKFTVSGIDKQKVGQIAANI
Rv0719|M.tuberculosis_H37Rv GSN-LEFALGYSHPVVIEAPEGITFAVQAPTKFTVSGIDKQKVGQIAANI
MLBr_01844|M.leprae_Br4923 GSN-LEFALGYSHPVVIEAPEGITFAVQSPTKFTITGIDKQKVGQISANI
TH_0308|M.thermoresistible__bu GSDTLEFSLGFSHPVVVKAPEGVTFNVESPTKFSISGIDKQKVGQISANI
Mflv_5032|M.gilvum_PYR-GCK GSN-LEFALGYSHPVLINAPEGVTFAVETPTKFSISGIDKQAVGQIAANI
Mvan_1337|M.vanbaalenii_PYR-1 GSN-LEFALGYSHPVLITAPEGVTFAVETPTKFSISGIDKQKVGQIAANI
MAB_3797c|M.abscessus_ATCC_199 GSN-LEFALGYSHPVLIEAPEGITFAVETPTKFSISGIDKQKVGQISANI
MSMEG_1470|M.smegmatis_MC2_155 GQN-LEFALGYSHPVLIEAPEGITFAVESPTKFSVSGIDKQKVGQISAVI
* : ***:**:****:: ****:** *::****:::***** ****:* *
MMAR_1050|M.marinum_M RRLRRPDPYKGKGVRYEGEQIRRKVGKTGK
MUL_0809|M.ulcerans_Agy99 RRLRRPDPYKGKGVRYEGEQIRRKVGKTGK
MAV_4450|M.avium_104 RRLRRPDPYKGKGVRYEGEQIRRKVGKTGK
Mb0740|M.bovis_AF2122/97 RRLRRPDPYKGKGVRYEGEQIRRKVGKTGK
Rv0719|M.tuberculosis_H37Rv RRLRRPDPYKGKGVRYEGEQIRRKVGKTGK
MLBr_01844|M.leprae_Br4923 RRLRRPDPYKGKGVRYEGEQIRRKVGKTGK
TH_0308|M.thermoresistible__bu RRLRRPDPYKGKGVRYEGEQIRRKVGKTGK
Mflv_5032|M.gilvum_PYR-GCK RRLRKSDPYKGKGIRYEGEQIRRKVGKTGK
Mvan_1337|M.vanbaalenii_PYR-1 RRLRKSDPYKGKGIRYEGEQIRRKVGKTGK
MAB_3797c|M.abscessus_ATCC_199 RRLRRPDPYKGKGIRYEGEQIRRKVGKTGK
MSMEG_1470|M.smegmatis_MC2_155 RRLRRPDPYKGKGVRYEGEQIRRKVGKTGK
****:.*******:****************