For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RNYYSAQQIREAEAPLLAALPDGALMRRAAYGLAAVIADELSRRAGVVAGRSVCGVVGSGDNGGDALWAL TFLRRRGVSASAVLLNPERAHRAGLAAFRKAGGRVVSVVPDDTDLVVDGVVGISGTGPLRPGAAAGIAEK ITAEGIPVIAVDIPSGVDVHTGAVDGPAFRAAVTVTFGGYKPVHALGDCGDVRLIDIGLELPESALREFD ASDVAAAWPVPGPADDKYTQGVTGVLAGSSTYPGAAVLCSGAAVAATSGMVRYAGTAAAEVVSHWPEVIA TQTEEAAGRVQAWVIGPGYGTAERQTRTLRRVLSTSLPVLVDADALTMLAEHPDLADLVASRPAATVLTP HAGEFARLAGGPPGPDRVDAARSLAVRLKATVLLKGNVTVIARPDGTAYVNRAHGSWAATAGSGDVLAGV IGALLSAGIAADKAAAMGAYVHARAAAAAAADPGPGKAPISASRLLGHLRWAIAEIG*
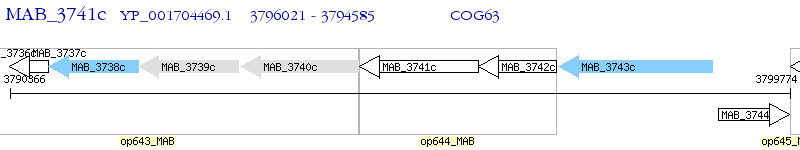
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3741c | - | - | 100% (478) | hypothetical protein MAB_3741c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3463c | - | 0.0 | 71.28% (477) | hypothetical protein Mb3463c |
| M. gilvum PYR-GCK | Mflv_4936 | - | 0.0 | 69.18% (477) | carbohydrate kinase, YjeF related protein |
| M. tuberculosis H37Rv | Rv3433c | - | 0.0 | 71.07% (477) | hypothetical protein Rv3433c |
| M. leprae Br4923 | MLBr_00373 | - | 0.0 | 69.18% (477) | hypothetical protein MLBr_00373 |
| M. marinum M | MMAR_1117 | - | 1e-180 | 68.97% (477) | transmembrane protein |
| M. avium 104 | MAV_4374 | - | 0.0 | 69.60% (477) | carbohydrate kinase |
| M. smegmatis MC2 155 | MSMEG_1573 | - | 1e-180 | 68.97% (477) | carbohydrate kinase family protein |
| M. thermoresistible (build 8) | TH_1696 | - | 1e-180 | 68.34% (477) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0877 | - | 1e-179 | 68.76% (477) | transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_1482 | - | 0.0 | 70.86% (477) | carbohydrate kinase, YjeF related protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1117|M.marinum_M MLAVMRHYYSVDAIRDAEAPLLASLPDGVLMRRAAFGLATEIGRELTRRS
MUL_0877|M.ulcerans_Agy99 ----MRHYYSVDAIRDAEAPLLASLPDGVLMRRAAFGLATEIGRELTRRS
Mb3463c|M.bovis_AF2122/97 ----MRHYYSVDTIRAAEAPLLASLPDGALMRRAAFGLATEIGRELTART
Rv3433c|M.tuberculosis_H37Rv ----MRHYYSVDTIRAAEAPLLASLPDGALMRRAAFGLATEIGRELTART
MAV_4374|M.avium_104 ----MRHYYSVDAIRAAEAPLLASLPDGALMRRAAFGLATEIAAELTART
MLBr_00373|M.leprae_Br4923 ----MRHYYSVAAIRDAEASLLASLPDGVLMKRAAYGLASVIIRELAVRT
MSMEG_1573|M.smegmatis_MC2_155 ----MRHYYSADAIREAEAPLLAALPDGALMRRAAYGLATVILRELSART
TH_1696|M.thermoresistible__bu ---VMLHYYSAEAIRAAEAPLLASLPTGALMRRAAFGLATVIGRELVART
Mflv_4936|M.gilvum_PYR-GCK -MSRMRYFYSADAIRDAEAPLLASLPDGVLMRRAAYGLATAIAAELG---
Mvan_1482|M.vanbaalenii_PYR-1 ----MRYFYSADAIRDAEAPVLASLPDGALMRRAAFGLATAIARELG---
MAB_3741c|M.abscessus_ATCC_199 ----MRNYYSAQQIREAEAPLLAALPDGALMRRAAYGLAAVIADELSRRA
* :**. ** ***.:**:** *.**:***:***: * **
MMAR_1117|M.marinum_M GGIFGRRVCAVVGSGDNGGDALWAATFLRRRGAAADAVLLNPQHTHPKAL
MUL_0877|M.ulcerans_Agy99 GGIFGRRVCAVVGSGDNGGDALWAATSLRRRGAAADAVLLNPQHTHPKAL
Mb3463c|M.bovis_AF2122/97 GGVVGRRVCAVVGSGDNGGDALWAATFLRRRGAAADAVLLNPDRTHRKAL
Rv3433c|M.tuberculosis_H37Rv GGVVGRRVCAVVGSGDNGGDALWAATFLRRRGAAADAVLLNPDRTHRKAL
MAV_4374|M.avium_104 GGVAGRRVCAVVGSGDNGGDALWAATFLRRRGAAADAILLNPERAHRKGL
MLBr_00373|M.leprae_Br4923 GGVTGRRVCAVVGSGDNGGDALWAATFLRRRGAAADAVLLNPDRVHRKAL
MSMEG_1573|M.smegmatis_MC2_155 GGVAGRRVCAVVGSGDNGGDALWAATFLRRRGAAADAVLLNPERTHATAL
TH_1696|M.thermoresistible__bu GGVAGRRVCAVVGSGDNGGDALWAGTFLRRRGASVSAILLAGDRTHPRAL
Mflv_4936|M.gilvum_PYR-GCK -TVSGKSVCAVVGSGDNGGDALWAATFLRRRGAGACAVLLDPERAHRKGL
Mvan_1482|M.vanbaalenii_PYR-1 -AVAGRTVCAVVGSGDNGGDALWAATFLRRRGAGASAVLLDPERAHRKAL
MAB_3741c|M.abscessus_ATCC_199 GVVAGRSVCGVVGSGDNGGDALWALTFLRRRGVSASAVLLNPERAHRAGL
: *: **.************** * *****... *:** ::.* .*
MMAR_1117|M.marinum_M AAFTKAGGRVVQNVSAATDLVIDGVVGISGSGPLRP-AAAEVFAAVDAAG
MUL_0877|M.ulcerans_Agy99 AAFTKAGGRVVQNVSAATDLVIDGVVGISGSGPLRR-AAAEVFAAVDAAG
Mb3463c|M.bovis_AF2122/97 AAFTKSGGRLVESVSAATDLVIDGVVGISGSGPLRP-AAAQVFAAVQAAA
Rv3433c|M.tuberculosis_H37Rv AAFTKSGGRLVESVSAATDLVIDGVVGISGSGPLRP-AAAQVFAAVQAAA
MAV_4374|M.avium_104 AAFGKAGGRIVESVSPTTDLVIDGVVGISGSGALRP-AAAEVFAAVDDAG
MLBr_00373|M.leprae_Br4923 VAFRKAGGRIVENVSAATDLVIDGVVGISGSGPLRP-AAAAVFATVSASG
MSMEG_1573|M.smegmatis_MC2_155 AAFRRAGGQIVQTVNPATDLVIDGVVGISGRGPLRP-NAAEVFAGTDAP-
TH_1696|M.thermoresistible__bu AAFRRAGGRVVETLPPDLDLVIDGVVGIGGSGPLRP-PAAAVFGAVDEAG
Mflv_4936|M.gilvum_PYR-GCK AAFRRAGGRIVETVPPTTDLVIDGVVGISGSGPLRP-HAAEIFAAVDVAG
Mvan_1482|M.vanbaalenii_PYR-1 AAFRRAGGRIVEMVPHTTDLVIDGVVGISGSGPLRP-GAAAVFDEVARAG
MAB_3741c|M.abscessus_ATCC_199 AAFRKAGGRVVSVVPDDTDLVVDGVVGISGTGPLRPGAAAGIAEKITAEG
.** ::**::*. : ***:******.* *.** ** :
MMAR_1117|M.marinum_M IPVVAVDIPSGIDVATGAITGPAVRAALTVTFGGLKPVHALADCGRVVLI
MUL_0877|M.ulcerans_Agy99 IPVVAVDIPSGIDVATGAITGPAVRAALTVTFGGLKPVHALADCGRVVLI
Mb3463c|M.bovis_AF2122/97 IPVVAVDIPSGIDVATGAITGPAVHAALTVTFGGLKPVHALADCGRVVLV
Rv3433c|M.tuberculosis_H37Rv IPVVAVDIPSGIDVATGAITGPAVHAALTVTFGGLKPVHALADCGRVVLV
MAV_4374|M.avium_104 IPVVAVDIPSGIDAATGATSGPAVHAVLTVTFGGLKPVHALGDCGRVKLI
MLBr_00373|M.leprae_Br4923 VPVVAVDLPSGIDVVTGVINGPAVHAALTVTFGGLKPVHALADCGDVTLV
MSMEG_1573|M.smegmatis_MC2_155 --VVAVDIPSGIDVHTGAAEGPHVRAALTVTFGGLKPVHALGECGRVELI
TH_1696|M.thermoresistible__bu IPVVAVDVPSGIDVHTGAAEGPHVRATVTVTFGGLKPVHALADCGRVDLI
Mflv_4936|M.gilvum_PYR-GCK IPVVAVDLPSGVDPYTGAADGAHVTPALTVTFGGFKPVHAFGDCGRLELV
Mvan_1482|M.vanbaalenii_PYR-1 IPVVAVDLPSGVDVHTGVADGPHVAPALTVTFGGLKPVHALGDCGRVELV
MAB_3741c|M.abscessus_ATCC_199 IPVIAVDIPSGVDVHTGAVDGPAFRAAVTVTFGGYKPVHALGDCGDVRLI
*:***:***:* **. *. . ..:****** *****:.:** : *:
MMAR_1117|M.marinum_M DIGLDLSDTEVLGLQAADVAAHWPVPGPRDDKYTQGVTGVLAGSSTYPGA
MUL_0877|M.ulcerans_Agy99 DIGLDLSDTEVLGLQAADVAAHWPVPGPRDDKYTQGVTGVLAGSSTYPGA
Mb3463c|M.bovis_AF2122/97 DIGLDLAHTDVLGFEATDVAARWPVPGPRDDKYTQGVTGVLAGSSTYPGA
Rv3433c|M.tuberculosis_H37Rv DIGLDLAHTDVLGFEATDVAARWPVPGPRDDKYTQGVTGVLAGSSTYPGA
MAV_4374|M.avium_104 DIGLDLPQTDVLGFEAADVAARWPVPGPHDDKYTQGVTGVMAGSATYPGA
MLBr_00373|M.leprae_Br4923 DIGLDLPDSDILGLQAADVAAYWPVPGVHDDKYTQGVTGVLAGSSTYPGA
MSMEG_1573|M.smegmatis_MC2_155 DIGLDLAPADLASFEAADVRALWPAPGPHDDKYTQGVTGVLSGSATYPGA
TH_1696|M.thermoresistible__bu DIGLDLEPGDLRGFDAAEVAARWPRPGPADDKYTQGVAGILAGSATYPGA
Mflv_4936|M.gilvum_PYR-GCK DIGLDLAPSDVGALDAGDVAACWPVPGPDDDKYTQGVTGVMAGSATYPGA
Mvan_1482|M.vanbaalenii_PYR-1 DIGLGLGHSDMLAFDAADVAARWPVPGPTDDKYTQGVTGVMAGSSTYPGA
MAB_3741c|M.abscessus_ATCC_199 DIGLELPESALREFDASDVAAAWPVPGPADDKYTQGVTGVLAGSSTYPGA
**** * : ::* :* * ** ** ********:*:::**:*****
MMAR_1117|M.marinum_M AVLCSGAAVAATSGMVRYAGSAAGEVLARWPEVIASPTPSTTGRVQAWVV
MUL_0877|M.ulcerans_Agy99 AVLCSGAAVAATSGMVRYAGSAAGEVLARWPEVIASPTPSTTGRVQAWVV
Mb3463c|M.bovis_AF2122/97 AVLCTGAAVAATSGMVRYAGTAHAEVLAHWPEVIASPTPAAAGRVQAWVV
Rv3433c|M.tuberculosis_H37Rv AVLCTGAAVAATSGMVRYAGTAHAEVLAHWPEVIASPTPAAAGRVQAWVV
MAV_4374|M.avium_104 AVLCTGAAVAATSGMVRYAGSAHREVLAHWPEVIASPTPASAGRVQSWVV
MLBr_00373|M.leprae_Br4923 AVLCTGAAVAATSGMVRYAGSAYTQVLAHWPEVIASATPTAAGRVQSWVV
MSMEG_1573|M.smegmatis_MC2_155 AVLSTGAAVAATSGMVRYAGSAAQQVLAYWPEVIASATPQDAGRVQAWVV
TH_1696|M.thermoresistible__bu AVLCVGAAVATTSGMVRYAGSAAAHVLSSWPEVIATERPETAGRVQAWVV
Mflv_4936|M.gilvum_PYR-GCK AILCTGAAVAATSGMVRYAGTAAGEVVSHWPEVIATSTPASAGRVQAWVV
Mvan_1482|M.vanbaalenii_PYR-1 AILCTGAAVAATSGMVRFAGTAAAEVVSHWPEVIATLGHATAGRVQAWAV
MAB_3741c|M.abscessus_ATCC_199 AVLCSGAAVAATSGMVRYAGTAAAEVVSHWPEVIATQTEEAAGRVQAWVI
*:*. *****:******:**:* .*:: ******: :****:*.:
MMAR_1117|M.marinum_M GPGLGTDEPGAAALWFALETNLPVIVDADGLTMLAAHP---ELVVNRSAP
MUL_0877|M.ulcerans_Agy99 GPGLGTDEPGAAALWFALETSLPVIVDADGLTMLAAHP---ELVVNRSAP
Mb3463c|M.bovis_AF2122/97 GPGLGTDEAGAAALWFALDTDLPVLVDADGLTMLADHP---DLVAGRNAP
Rv3433c|M.tuberculosis_H37Rv GPGLGTDEAGAAALWFALDTDLPVLVDADGLTMLADHP---DLVAGRNAP
MAV_4374|M.avium_104 GPGLGTDDAGAAALWFALETDLPVIVDADGLTMLAAHP---ELVANRAAP
MLBr_00373|M.leprae_Br4923 GPGLGIDATATAALWFALETDLPVLVDADGLTMLAAHP---DLVINRNAP
MSMEG_1573|M.smegmatis_MC2_155 GPGLGTDDTAKAALLFALETDLPVIVDADALTMLAADP---APVTGRSAP
TH_1696|M.thermoresistible__bu GPGLGTDDTAAETLEFALRTDLPVLVDADGLTLLAARP---ELVRGRSAP
Mflv_4936|M.gilvum_PYR-GCK GPGLGTDEVAAAALVHALETDLPVIVDADGLTILAAHP---EIVQGRTAP
Mvan_1482|M.vanbaalenii_PYR-1 GPGLGTDETAAAALVFALETDLPVIVDADGLTILSAHP---ELVADRTAP
MAB_3741c|M.abscessus_ATCC_199 GPGYGTAERQTRTLRRVLSTSLPVLVDADALTMLAEHPDLADLVASRPAA
*** * :* .* *.***:****.**:*: * * .* *.
MMAR_1117|M.marinum_M TVLTPHAGEFARLAGSGPGDDRVGATRKLADAFGATVLLKGNVTVIADPG
MUL_0877|M.ulcerans_Agy99 TVLTPHAGEFARLAGSGPGDDRVGATRKLADAFGATVLLKGNVTVIADPG
Mb3463c|M.bovis_AF2122/97 TVLTPHAGEFARLAGAPPGDDRVGACRQLADALGATVLLKGNVTVIADPG
Rv3433c|M.tuberculosis_H37Rv TVLTPHAGEFARLAGAPPGDDRVGACRQLADALGATVLLKGNVTVIADPG
MAV_4374|M.avium_104 TVLTPHAGEFARLAGSPPGDDRVGATRKLADTLGVTVLLKGNVTVIADPG
MLBr_00373|M.leprae_Br4923 TVLTPHASEFARLAGTPPGDDRVGACRKLADSFGATVLLKGNVTVIADPG
MSMEG_1573|M.smegmatis_MC2_155 TVLTPHAGEFARLAGAPPGTDRVAAARELAERLGVTLLLKGNVTVIATPG
TH_1696|M.thermoresistible__bu TVLTPHAGEFARLASGPPGADRVAAARRLAEDLGATVLLKGHVTVIAEPG
Mflv_4936|M.gilvum_PYR-GCK TVLTPHAGEFARLAGSSPGAERVADTRRLADRFGATVLLKGNVTVVAEPD
Mvan_1482|M.vanbaalenii_PYR-1 TVLTPHAGEFARLAGAPPGDDRVAATRRLADRLGVTVLLKGNVTVVAEPG
MAB_3741c|M.abscessus_ATCC_199 TVLTPHAGEFARLAGGPPGPDRVDAARSLAVRLKATVLLKGNVTVIARPD
*******.******. ** :** * ** : .*:****:***:* *.
MMAR_1117|M.marinum_M GRVYLSPAGQSWAATAGSGDVLSGMIGALLAAGLAPAEAAAAAAFVHARA
MUL_0877|M.ulcerans_Agy99 GRVYLSPAGQSWAATAGSGDVLSGMIGALLAAGLAPAEAAAAAAFVHARA
Mb3463c|M.bovis_AF2122/97 GPVYLNPAGQSWAATAGSGDVLSGMIGALLASGLPSGEAAAAAAFVHARA
Rv3433c|M.tuberculosis_H37Rv GPVYLNPAGQSWAATAGSGDVLSGMIGALLASGLPSGEAAAAAAFVHARA
MAV_4374|M.avium_104 GPVYLNPAGQSWAATAGSGDVLSGMIGALLASGLPAGEAAAAAAFVHARA
MLBr_00373|M.leprae_Br4923 GPVYLNPAGQSWAATAGSGDVLSGMIGALLAAGLPAAEAAAAAAFVHARA
MSMEG_1573|M.smegmatis_MC2_155 ATTYLNPAGQSWAATAGSGDVLSGIIGALLASGLPPGEAAAAAAFVHARA
TH_1696|M.thermoresistible__bu GPVYLNPAGESWSATAGSGDVLSGMIGALLASGLPAVEAAAAAAFVHSRA
Mflv_4936|M.gilvum_PYR-GCK GQVYLNAAGNSWAATAGSGDVLSGMIGALLAAGLPTGQAAAAAAYVHSRA
Mvan_1482|M.vanbaalenii_PYR-1 GPVRLNAAGNSWAATAGSGDVLSGVIGALLAAGLPPGEAAAAAAFVHARA
MAB_3741c|M.abscessus_ATCC_199 GTAYVNRAHGSWAATAGSGDVLAGVIGALLSAGIAADKAAAMGAYVHARA
. . :. * **:*********:*:*****::*:.. :*** .*:**:**
MMAR_1117|M.marinum_M AELSALDPGPDGAPTSASRILHHIRAALAAL-
MUL_0877|M.ulcerans_Agy99 AELSALDPGPDGAPTSASRILRHIRAALAAL-
Mb3463c|M.bovis_AF2122/97 AAAAAADPGPGDAPTSASRISGHIRAALAAL-
Rv3433c|M.tuberculosis_H37Rv SAAAAADPGPGDAPTSASRISGHIRAALAAL-
MAV_4374|M.avium_104 AASSAADPGPGEAPTSSSRMVPHIRGALAAL-
MLBr_00373|M.leprae_Br4923 AALSAADPGPGDVPTSASRMVSHIRTALAAL-
MSMEG_1573|M.smegmatis_MC2_155 ANLSAADAGPRPVPTSASRILVHVRAAIAAL-
TH_1696|M.thermoresistible__bu ANLSAADPGPHSTQTSASRILQHIRTAVARL-
Mflv_4936|M.gilvum_PYR-GCK ADLSARDPGPAAAPTSASRILAHLRTALASIL
Mvan_1482|M.vanbaalenii_PYR-1 ADLSARDPGPAAAPTSASRILAHLRPALASIL
MAB_3741c|M.abscessus_ATCC_199 AAAAAADPGPGKAPISASRLLGHLRWAIAEIG
: :* *.** . *:**: *:* *:* :