For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LDGRLNPHAPPGDGGDGPAWSRKRQPYEPPPRERGRSVVPYAELHAHSAFSFLDGASLPEEMVQEAARLD LKALAITDHDGFYGVVRFAEAAKELGLPTVFGAELSLGGQGNTEDSVHLLVLARGQEGYRRLSRQMAGAH LSGGTPKDRKGKPRYDLDALTEAAGGHWHILTGCRKGHVRRALAGGGPVAAERALANLVDRFGADRVSIE LNRHGHPGEDERNAELAALARGFGVGVIATTAAHFATPESGRLAMAMAAVRARKSLDDAAGWLDPVGGAH LRSGDEMARLFSHYPEVVTAAAELGEECAFDLRLIAPQLPPFDVPSGHTEDSWLRHLALEGAARRYGPRA GAQKAYAQIERELEIIAQLNFPGYFLVVHDITQFCRRSDILCQGRGSAANSAVCYALGVTNVDPVDNGLL FERFLSPARDGPPDIDIDIESDEREQAIQYVYNKYGREYAAQVANVITYRGRMAVRDMAKALGFAQGQQD AWSKQMGHWGGLADAAAVDGIPPQVIQLARQIKDFPRHMGIHSGGMVICDRPLADVVPVEWARMENRSVL QWDKDDCAAVGLVKFDLLGLGMLSALHYCIDLVREHKGIDVDLAHINLKEQAVYEMLARADSVGVFQVES RAQMATLPRLKPKCFYDLVVEVALIRPGPIQGGSVHPYIRRYNKIDKDWQHDHPSMAAALDKTLGVPLFQ EQLMQLAVDVAGFSPAESDQLRRAMGSKRSPERMERLRNRFYEGMRNLHGITGELADRIYEKLYAFANFG FPESHAQSFASLVFYSSWFKLHHPAAFCAALLRAQPMGFYSPQSLVADARRHGVTVHGPDVNASLSYATL ENAGLEVRIGLGAVRHIGDDLAQAIVEERKVRGPFVSLLDLTGRIQLTTAQVEALATGGALGCFDMSRRE ALWVAGAAAAQRPDRLPGVGVSSRIPALPEMGEVTRAAADVWATGVSPDSYPTQFLRARLDAMGVVPAKG LFGVPDGSRVLVAGAVTHRQRPATAAGVTFINLEDETGMVNVVCSVGLWARYRKLAVTARALIIRGQVQN ASGAVSVVADQLRPLDLQIRSTSRDFR*
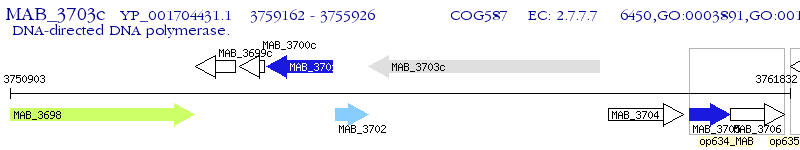
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3703c | dnaE2 | - | 100% (1078) | error-prone DNA polymerase |
| M. abscessus ATCC 19977 | MAB_2696c | dnaE | e-117 | 28.83% (1103) | DNA polymerase III subunit alpha |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3405c | dnaE2 | 0.0 | 76.82% (1083) | error-prone DNA polymerase |
| M. gilvum PYR-GCK | Mflv_4893 | dnaE2 | 0.0 | 76.02% (1076) | error-prone DNA polymerase |
| M. tuberculosis H37Rv | Rv3370c | dnaE2 | 0.0 | 76.82% (1083) | error-prone DNA polymerase |
| M. leprae Br4923 | MLBr_01207 | dnaE | 1e-112 | 28.05% (1105) | DNA polymerase III subunit alpha |
| M. marinum M | MMAR_1158 | dnaE2 | 0.0 | 75.98% (1095) | DNA polymerase III, alpha chain, DnaE2 |
| M. avium 104 | MAV_4335 | dnaE2 | 0.0 | 77.89% (1072) | error-prone DNA polymerase |
| M. smegmatis MC2 155 | MSMEG_1633 | dnaE2 | 0.0 | 76.58% (1076) | error-prone DNA polymerase |
| M. thermoresistible (build 8) | TH_0581 | - | 1e-118 | 28.92% (1117) | DNA polymerase III alpha subunit |
| M. ulcerans Agy99 | MUL_0923 | dnaE2 | 0.0 | 75.71% (1095) | error-prone DNA polymerase |
| M. vanbaalenii PYR-1 | Mvan_1530 | dnaE2 | 0.0 | 76.38% (1071) | error-prone DNA polymerase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3405c|M.bovis_AF2122/97 MGWSNGPPSWAEMERVLNGKPRHAGVP-----AFDADGDVPRSRKRGAYQ
Rv3370c|M.tuberculosis_H37Rv MGWSNGPPSWAEMERVLNGKPRHAGVP-----AFDADGDVPRSRKRGAYQ
MMAR_1158|M.marinum_M MGWGNGPPSWAEMERVLDGKPRHAGVPDAAGPTAEAGWDGPLSRKRETYA
MUL_0923|M.ulcerans_Agy99 ------------MERVLDGKPRHAGVPDAAGPTAEAGWDGPLSRKRETYA
MAV_4335|M.avium_104 MGWFNGPPSWAEMERVLDSKPRRAGES------PAPEPDGPLSRGRATYR
Mflv_4893|M.gilvum_PYR-GCK ------------MRRVLEGKPRRAGWP-IDAQVGDGGDSPAWSSKRGQYR
Mvan_1530|M.vanbaalenii_PYR-1 MGWHTGPPSWTEMERVLSGKPRRAGWP-IDQQIGDGGDSPAWSRKRGEYH
MSMEG_1633|M.smegmatis_MC2_155 MGWHNGPPSWSEMERVLTSKPRRSGLP-LESP-GDGGDSPAWSRKRGAYE
MAB_3703c|M.abscessus_ATCC_199 ---------------MLDGRLNPHAPP------GDGGDGPAWSRKRQPYE
MLBr_01207|M.leprae_Br4923 --------------------------------------------------
TH_0581|M.thermoresistible__bu --------------------------------------------------
Mb3405c|M.bovis_AF2122/97 P-PGRE-RVGSSVAYAELHAHSAYSFLDGASTPEELVEEAARLGLCALAL
Rv3370c|M.tuberculosis_H37Rv P-PGRE-RVGSSVAYAELHAHSAYSFLDGASTPEELVEEAARLGLCALAL
MMAR_1158|M.marinum_M PKPDAN-RVDSSIAYAELHAHSAYSFLDGASTPEELVEEAARLDLRALAL
MUL_0923|M.ulcerans_Agy99 PKPDAN-RVDSSIAYAELHAHSAYSFLDGASTPEELVEEAARLDLRALAL
MAV_4335|M.avium_104 PPDEGR-AARSSVPYAELHAHSAFSFLDGASTPEEMVEEAARLDLRALAL
Mflv_4893|M.gilvum_PYR-GCK APESRGPATGRSMPYAELHAHSAYSFLDGASTPEELVEEASRLGLRALAL
Mvan_1530|M.vanbaalenii_PYR-1 APEGPG-AQESSTPYAELHAHSAYSFLDGASTPEELVEEAARLNLRAIAL
MSMEG_1633|M.smegmatis_MC2_155 PPDQAR-MPASALPYAELHAHSAYSFLDGASTPEELVEEAARLNLRAIAL
MAB_3703c|M.abscessus_ATCC_199 PPPRER--GRSVVPYAELHAHSAFSFLDGASLPEEMVQEAARLDLKALAI
MLBr_01207|M.leprae_Br4923 ---------MNQSSFVHLHNHTEYSMLDGAAKITPMFAEVERLQMPAVGM
TH_0581|M.thermoresistible__bu ----------MSSSFVHLHNHTEFSMLDGAAKIGPLLAEAQRLEMSAIGM
.:..** *: :*:****: :. *. ** : *:.:
Mb3405c|M.bovis_AF2122/97 TDHDGLYGAVRFAEAAAELDVRTVFGAELSLGATARTERPDPPG------
Rv3370c|M.tuberculosis_H37Rv TDHDGLYGAVRFAEAAAELDVRTVFGAELSLGATARTERPDPPG------
MMAR_1158|M.marinum_M TDHNGLYGAVRFAEAASGLGVRTVFGAELSLGPEARTEQPDPPG------
MUL_0923|M.ulcerans_Agy99 TDHNGLYGAVRFAEAASGLGVRTVFGAELSLGPEARTEQPDPPG------
MAV_4335|M.avium_104 TDHDGLYGAVRFAEAAAELDVRTVFGAELSLGPSARTEAPDPPG------
Mflv_4893|M.gilvum_PYR-GCK TDHDGLYGVVRFAEAAKELDVDTVFGAELSLGGGTRTDVPDPPG------
Mvan_1530|M.vanbaalenii_PYR-1 TDHDGLYGVVRFAEAARELDVATVFGAELSLGGGTRTDVPDPPG------
MSMEG_1633|M.smegmatis_MC2_155 TDHDGLYGVVRFAEAARELDVATVFGAELSLSNVARTEDPDPPG------
MAB_3703c|M.abscessus_ATCC_199 TDHDGFYGVVRFAEAAKELGLPTVFGAELSLGGQGNTEDS----------
MLBr_01207|M.leprae_Br4923 TDHGNMFGASEFYNTAIKAGIKPIIGVEAYIAPGSRFDTRRITWGDPSQK
TH_0581|M.thermoresistible__bu TDHGNMFGASEFYNAATDAGIKPIIGVEAYIAPGSRFDTKRVQWGDPSQK
***..::*. .* ::* .: .::*.* :. . :
Mb3405c|M.bovis_AF2122/97 ----------PHLLVLARGPEGYRRLSRQLAAAHLAGGE----KGKPRYD
Rv3370c|M.tuberculosis_H37Rv ----------PHLLVLARGPEGYRRLSRQLAAAHLAGGE----KGKPRYD
MMAR_1158|M.marinum_M ----------PHLLVLARGPEGYRRLSRQLAAAHLAGGE----KGKPRYD
MUL_0923|M.ulcerans_Agy99 ----------PHLLVLARGPEGYRRLSRQLAAAHLAGGE----KGKPRYD
MAV_4335|M.avium_104 ----------PHLLVLARGPEGYRRLSRQLAAAHLAGGE----KGKPRYD
Mflv_4893|M.gilvum_PYR-GCK ----------PHLLVLARGPEGYRRLSRQLAAAHLAGGE----KGVLRYD
Mvan_1530|M.vanbaalenii_PYR-1 ----------PHLLVLARGPEGYRRLSRQLAAAHLAGGE----KGVLRYD
MSMEG_1633|M.smegmatis_MC2_155 ----------PHLLVLARGPEGYRRLSREIAKAHLAGGE----KGRPRYD
MAB_3703c|M.abscessus_ATCC_199 ----------VHLLVLARGQEGYRRLSRQMAGAHLSGGTPKDRKGKPRYD
MLBr_01207|M.leprae_Br4923 ADDVSAGGAYTHLTMVAENAAGLRNLFKLSSLASFEGQLS----KWSRMD
TH_0581|M.thermoresistible__bu SDDVSGSGAYTHMTMVAETADGLRNMFKLSSLASFEGQLG----KWSRMD
*: ::*. * *.: : : * : * * *
Mb3405c|M.bovis_AF2122/97 FDALTEAAGGHWHILTGCRKGHVRQALSQGGPAAAQRALADLVDRFTPSR
Rv3370c|M.tuberculosis_H37Rv FDALTEAAGGHWHILTGCRKGHVRQALSQGGPAAAQRALADLVDRFTPSR
MMAR_1158|M.marinum_M FDSLTEAAGGHWHILTGCRKGHVRQALRQGGPDAAERALADLVDRFTAGR
MUL_0923|M.ulcerans_Agy99 FDSLTEAAGGHWHILTGCRKGHVRQALRQGGPDAAERVLADLVDRFTAGR
MAV_4335|M.avium_104 LDALTEAAGGHWHILTGCRKGHVRQALSEGGPDEAARALADLVDRFGAAR
Mflv_4893|M.gilvum_PYR-GCK FDALTEAAGGHWQILTGCRKGHVRQALLRGGDSAAEAALADLVDRFGRER
Mvan_1530|M.vanbaalenii_PYR-1 FDALTEAAGGHWQILTGCRKGHVRQALSTGGPEAAEAALADLVDRFGPDR
MSMEG_1633|M.smegmatis_MC2_155 FDQLTEAAGGHWHILTGCRKGHVRQALSDGGPDAAAAALADLVDRFGADR
MAB_3703c|M.abscessus_ATCC_199 LDALTEAAGGHWHILTGCRKGHVRRALAGGGPVAAERALANLVDRFGADR
MLBr_01207|M.leprae_Br4923 AELIGEYAEG-IIVTTGCPSGEVQTRLRLGHDREALESAAKWREIVGPDN
TH_0581|M.thermoresistible__bu AELIAEHATG-IIATTGCPSGEVQTRLRLGHHREALEAAAKWRDIFGPDN
: : * * * *** .*.*: * * * *. : . .
Mb3405c|M.bovis_AF2122/97 VSIELTHHGHPLDDERNAALAGLAPRFGVGIVATTGAHFADPSRGRLAMA
Rv3370c|M.tuberculosis_H37Rv VSIELTHHGHPLDDERNAALAGLAPRFGVGIVATTGAHFADPSRGRLAMA
MMAR_1158|M.marinum_M VSVELTHHGHPLDDENNAMLAGLAPRFGVGVVATTGAHFAHPARSRLAMA
MUL_0923|M.ulcerans_Agy99 VSVELTHHGHPLDDENNAMLAGLAPRFGVGVVATTGAHFAHPARSRLAMA
MAV_4335|M.avium_104 VSIELTRHGQPLDDERNAALAALAPRFGVGVVATTGAHFAGPSRRRLAMA
Mflv_4893|M.gilvum_PYR-GCK VTVELTHHGHPLDDERNAALAALAPRFGLTVIATTAAHFAEPSRGRLAMA
Mvan_1530|M.vanbaalenii_PYR-1 VTVELTHHGHPLDDERNAALAALAPRFGLGVVATTAAHFAEPARGRLAMA
MSMEG_1633|M.smegmatis_MC2_155 VSIELTHHGHPCDDERNAALAELAPRFGLGVVATTAAHFATPSRGRLAMA
MAB_3703c|M.abscessus_ATCC_199 VSIELNRHGHPGEDERNAELAALARGFGVGVIATTAAHFATPESGRLAMA
MLBr_01207|M.leprae_Br4923 YFLELMDHGLSIEQRVREGLLNIGRKLNIPPLATNDCHYVTRDAVHNHEA
TH_0581|M.thermoresistible__bu YFLELMDHGLDIERRVRDGLLEIGRKLNIPPLATNDCHYVTRDAAQNHEA
:** ** : . . * :. :.: :**. .*:. : *
Mb3405c|M.bovis_AF2122/97 MAAIRARRSLDSAAGWLAPLGGAHLRSGEEMARLFAWCPEAVTAAAELGE
Rv3370c|M.tuberculosis_H37Rv MAAIRARRSLDSAAGWLAPLGGAHLRSGEEMARLFAWCPEAVTAAAELGE
MMAR_1158|M.marinum_M MGAIRARQSLDSAAGWLAPLGGSHLRSGAEMARLFAQRPDVVTAAAELGE
MUL_0923|M.ulcerans_Agy99 MGAIRARQSLDSAAGWLAPLGGSHLRSGAEMARLFAQRPDVVTAAAELGE
MAV_4335|M.avium_104 VGAIRARESLDSAAGWLAPLGGSHLRSGEEMARLFAWCPEAVTAAAELGE
Mflv_4893|M.gilvum_PYR-GCK MGAIRARNSIDEAAGYLAPLGGSHLRSGEEMARVFAHCPEVVTAAADLGE
Mvan_1530|M.vanbaalenii_PYR-1 MGAIRARNSIDEAAGYLAPLGGSHLRSGDEMARMFAHCPEVVTAAAELGE
MSMEG_1633|M.smegmatis_MC2_155 MAAIRARNSIDTAAGWLAPLGGVHLRSGEEMARLFD--PEFVAAAADLGE
MAB_3703c|M.abscessus_ATCC_199 MAAVRARKSLDDAAGWLDPVGGAHLRSGDEMARLFSHYPEVVTAAAELGE
MLBr_01207|M.leprae_Br4923 LLCVQTGKTLSDPNRFKFHGDGYYLKSAAEMRQLWDDEVPGACDSTLLIA
TH_0581|M.thermoresistible__bu LLCVQTGKTLSDPNRFKFDGDGYYLKSAAEMRALWDDEVPGACDNTLLIA
: .::: .::. . : .* :*:*. ** :: . : *
Mb3405c|M.bovis_AF2122/97 RCAFGLQLIAP---RLPPFDVPDGHTEDSWLRSLVMAGARERYGPPKSAP
Rv3370c|M.tuberculosis_H37Rv RCAFGLQLIAP---RLPPFDVPDGHTEDSWLRSLVMAGARERYGPPKSAP
MMAR_1158|M.marinum_M QCAFGLALIAP---RLPPFDVPDGHTEDSWLRSLVMSGARERYGSTERSP
MUL_0923|M.ulcerans_Agy99 QCAFGLALIAP---RLPPFDVPDGHTEDSWLRSLVMSGARERYGSTERSP
MAV_4335|M.avium_104 QCAFGLALIAP---RLPPFDVPDGHTEDSWLRQLTMTGARDRYGSPEHAP
Mflv_4893|M.gilvum_PYR-GCK QCAFGLALIAP---QLPPFEVPAGHTENSWLRHLVMQGARERYGPPERAS
Mvan_1530|M.vanbaalenii_PYR-1 QCAFGLALIAP---QLPPFDVPDGHTESSWLRHLVMQGARERYGPPERAS
MSMEG_1633|M.smegmatis_MC2_155 QCAFGLALIAP---QLPPFDVPDGHTEDSWLRHLAMAGAARRYGPPERAP
MAB_3703c|M.abscessus_ATCC_199 ECAFDLRLIAP---QLPPFDVPSGHTEDSWLRHLALEGAARRYGPRAGAQ
MLBr_01207|M.leprae_Br4923 ERVQSYADVWEPRNRMPVFPVPVGHDQASWLRHEVDAGLKRRFPDGPPNG
TH_0581|M.thermoresistible__bu ERVQSYADVWTPRDRMPVFPVPEGHTQESWLRHEVRAGLQRRFPDGVPQE
. . . : ::* * ** ** : **** . * *:
Mb3405c|M.bovis_AF2122/97 ---RAYSQIEHELKVIAQLRFPGYFLVVHDITRFCRDNDILC-QGRGSAA
Rv3370c|M.tuberculosis_H37Rv ---RAYSQIEHELKVIAQLRFPGYFLVVHDITRFCRDNDILC-QGRGSAA
MMAR_1158|M.marinum_M RARKAYTQIEHELKVIAQLSFPGYFLVVHDITQFCRRNDILC-QGRGSAA
MUL_0923|M.ulcerans_Agy99 RARKAYAQIEHELKVIAQLSFPGYFLVVHDITQFCRRNDILC-QGRGSAA
MAV_4335|M.avium_104 ---RAYAQIEHELKVIAQLQFPGYFLVVHDIARFCRENNILC-QGRGSAA
Mflv_4893|M.gilvum_PYR-GCK ---RAYAQIEHELRVIEQLNFPGYFLVVHDITRFCRENAILS-QGRGSAA
Mvan_1530|M.vanbaalenii_PYR-1 ---RAYAQIEHELAVIEQLNFPGYFLVVHDITRFCRDNNILS-QGRGSAA
MSMEG_1633|M.smegmatis_MC2_155 ---KAYAQIEHELRIIEQLRFPGYFLVVHDITQFCRDNNILC-QGRGSAA
MAB_3703c|M.abscessus_ATCC_199 ---KAYAQIERELEIIAQLNFPGYFLVVHDITQFCRRSDILC-QGRGSAA
MLBr_01207|M.leprae_Br4923 ----YVERAAYEIDVICDKGFPAYFLIVADLVNHARSVGIRVGPGRGSAA
TH_0581|M.thermoresistible__bu ----YLDRAEYEIEVICAKGYPSYFLIVADLVNYARSVDIRVGPGRGSAA
: *: :* :*.***:* *:....* * ******
Mb3405c|M.bovis_AF2122/97 NSAVCYALGVTAVDPVANELLFERFLSPARDGPPDIDIDIESDQREKVIQ
Rv3370c|M.tuberculosis_H37Rv NSAVCYALGVTAVDPVANELLFERFLSPARDGPPDIDIDIESDQREKVIQ
MMAR_1158|M.marinum_M NSAVCYALGVTAVDPIANELLFERFLSPERDGPPDIDIDIESDQREKVIQ
MUL_0923|M.ulcerans_Agy99 NSAVCYALGVTAVDPIANELLFERFLSPERDGPPDIDIDIESDQREKVIQ
MAV_4335|M.avium_104 NSAVCYALGVTAVDPVANELLFERFLSPARDGPPDIDMDIESDQREKVIQ
Mflv_4893|M.gilvum_PYR-GCK NSAVCYALKVTNVDPIANGLLFERFLSPARDGPPDIDIDIESDLREKAIQ
Mvan_1530|M.vanbaalenii_PYR-1 NSAVCYALKVTNVDPIANDLLFERFLSPARDGPPDIDIDIESDLRENAIQ
MSMEG_1633|M.smegmatis_MC2_155 NSAVCYALGVTNVDPIANDLLFERFLSPARDGPPDIDIDIESDLRENVIQ
MAB_3703c|M.abscessus_ATCC_199 NSAVCYALGVTNVDPVDNGLLFERFLSPARDGPPDIDIDIESDEREQAIQ
MLBr_01207|M.leprae_Br4923 GSLAAYALGITDIDPIPHGLLFERFLNPERTSMPDIDIDFDDRRRGEMVR
TH_0581|M.thermoresistible__bu GSLVAYALRITDIDPIEHGLLFERFLNPERASMPDIDIDFDDRRRGEMVR
.* ..*** :* :**: : *******.* * . ****:*::. * : ::
Mb3405c|M.bovis_AF2122/97 YVYHKYGRDYAAQVANVITYRGRSAVRDMARALGFSP-------------
Rv3370c|M.tuberculosis_H37Rv YVYHKYGRDYAAQVANVITYRGRSAVRDMARALGFSP-------------
MMAR_1158|M.marinum_M YVYNKYGRDYAAQVANVITYRGRSAVRDMARALGFSQ-------------
MUL_0923|M.ulcerans_Agy99 YVYNKYGRDYAAQVANVITYRGRSAVRDMARALGFSQ-------------
MAV_4335|M.avium_104 YVYDRYGRDYAAQVANVITYRGKIAVRDMARALGFSQ-------------
Mflv_4893|M.gilvum_PYR-GCK YVYERYGREYAAQVANVITYRGRSAVRDMARALGFSQ-------------
Mvan_1530|M.vanbaalenii_PYR-1 YVYQRYGREYAAQVANVITYRGRSAVRDMARALGFSQ-------------
MSMEG_1633|M.smegmatis_MC2_155 YVYERYGRDYAAQVANVITYRGRSAIRDMARALGFSQ-------------
MAB_3703c|M.abscessus_ATCC_199 YVYNKYGREYAAQVANVITYRGRMAVRDMAKALGFAQ-------------
MLBr_01207|M.leprae_Br4923 YAADKWGHDRVAQVITFGTIKTKAALKDSARIHYGQPGFAMADRITKALP
TH_0581|M.thermoresistible__bu YAAEKWGHERVAQVITFGTIKTKAALKDAARVHYGQPGFAIADRITKALP
*. .::*:: .*** .. * : : *::* *:
Mb3405c|M.bovis_AF2122/97 -----------GQQDAWSKQVSHWTGQAD--DVDGIPEQVIDLATQIRNL
Rv3370c|M.tuberculosis_H37Rv -----------GQQDAWSKQVSHWTGQAD--DVDGIPEQVIDLATQIRNL
MMAR_1158|M.marinum_M -----------GQQDAWSKQIGHWNATPD--DVEGIPEQVIDLAAQIRNL
MUL_0923|M.ulcerans_Agy99 -----------GQQDAWSKQIGHWNATLD--DVEGIPEQVIDLAAQIRNL
MAV_4335|M.avium_104 -----------GQQDAWSKQISSWSGLADSPDVEGIPPQVIDLANQVRNL
Mflv_4893|M.gilvum_PYR-GCK -----------GQQDAWSKQISQWGNLADATHVEDIPEPVIDLAMQISHL
Mvan_1530|M.vanbaalenii_PYR-1 -----------GQQDAWSKQVSQWGNLADATHVEDIPGPVVDLAKQISNL
MSMEG_1633|M.smegmatis_MC2_155 -----------GQQDAWSKHLSRWDGRPDSPDVAEIPEQVIELANQIANL
MAB_3703c|M.abscessus_ATCC_199 -----------GQQDAWSKQMGHWGGLADAAAVDGIPPQVIQLARQIKDF
MLBr_01207|M.leprae_Br4923 PAIMAKDIPLSGITDPAHERFKEAAEVRSLIETDSDVRIIYQTARGLEGL
TH_0581|M.thermoresistible__bu PPIMAKDIPLSGITDPNHERYKEAAEVRGLIESDPDVRRIYETALGLEGL
* *. :: . : : * : :
Mb3405c|M.bovis_AF2122/97 PRHLGIHSGGMVICDRPIADVCPVEWARMANRSVLQWDKDDCAAIGLVKF
Rv3370c|M.tuberculosis_H37Rv PRHLGIHSGGMVICDRPIADVCPVEWARMANRSVLQWDKDDCAAIGLVKF
MMAR_1158|M.marinum_M PRHMGIHSGGMVICDRPIADVCPVEWARMADRSVLQWDKDDCAAIGLVKF
MUL_0923|M.ulcerans_Agy99 PRHMGIHSGGMVICDRPIADVCPVEWARMADRSVLQWDKDDCAAIGLVKF
MAV_4335|M.avium_104 PRHLGIHSGGMVICDRPIADVCPVEWARMENRSVLQWDKDDCAAIGLVKF
Mflv_4893|M.gilvum_PYR-GCK PRHMGIHSGGMVICDRPIADVCPVEWARMENRSVLQWDKDDCAAIGLVKF
Mvan_1530|M.vanbaalenii_PYR-1 PRHMGIHSGGMVICDRPIADVCPVEWARMENRSVLQWDKDDCAAIGLVKF
MSMEG_1633|M.smegmatis_MC2_155 PRHMGIHSGGMVICDRPIADVCPVEWARMENRSVLQWDKDDCAAIGLVKF
MAB_3703c|M.abscessus_ATCC_199 PRHMGIHSGGMVICDRPLADVVPVEWARMENRSVLQWDKDDCAAVGLVKF
MLBr_01207|M.leprae_Br4923 IRNAGVHACAVILSSEPLTEAIPLWKRPQDGAIITGWDYPACEAIGLLKM
TH_0581|M.thermoresistible__bu VRNAGVHACAVIMSSEPLIDAIPLWKRPQDGAIITGWDYPACEAIGLLKM
*: *:*: .:::...*: :. *: . : ** * *:**:*:
Mb3405c|M.bovis_AF2122/97 DLLGLGMLSALHYAKDLVAEHKGIEVDLARLDLSEPAVYEMLARADSVGV
Rv3370c|M.tuberculosis_H37Rv DLLGLGMLSALHYAKDLVAEHKGIEVDLARLDLSEPAVYEMLARADSVGV
MMAR_1158|M.marinum_M DLLGLGMLSALHYARDLVAEHKGIEVDLARLDLSEPAVYEMLARADSVGV
MUL_0923|M.ulcerans_Agy99 DLLGLGMLSALHYARDLVAEHKGIEVDLARLDLSEPAVYEMLARADSVGV
MAV_4335|M.avium_104 DLLGLGMLSALHYAIDLVAEHKGIEVDLARLDLSEPAVYEMLARADSVGV
Mflv_4893|M.gilvum_PYR-GCK DLLGLGMLSALHYAIDLVAEHKGLEVDLAKLDLSEPAVYEMLQRADSVGV
Mvan_1530|M.vanbaalenii_PYR-1 DLLGLGMLSALHYAIDLVAEHKGIEVDLAKLDLSEPAVYEMLARADSVGV
MSMEG_1633|M.smegmatis_MC2_155 DMLGLGMLSALHYAIDLVAEHKGIEVDLATLDLSEPAVYEMLQRADSVGV
MAB_3703c|M.abscessus_ATCC_199 DLLGLGMLSALHYCIDLVREHKGIDVDLAHINLKEQAVYEMLARADSVGV
MLBr_01207|M.leprae_Br4923 DFLGLRNLTIIGDAIENIKTNRGIDLDLESVPLDDQATYELLGRGDTLGV
TH_0581|M.thermoresistible__bu DFLGLRNLTIIGDALENIKANRGIDLDLDTIPMDDPATYELLGRGDTLGV
*:*** *: : . : : ::*:::** : :.: *.**:* *.*::**
Mb3405c|M.bovis_AF2122/97 FQVESRAQMATLPRLKPRVFYDLVVEVALIRPGPIQGGSVHPYIRRRNGV
Rv3370c|M.tuberculosis_H37Rv FQVESRAQMATLPRLKPRVFYDLVVEVALIRPGPIQGGSVHPYIRRRNGV
MMAR_1158|M.marinum_M FQVESRAQMATLPRLKPRVFYDLVVEVALIRPGPIQGGSVHPYIRRRNGI
MUL_0923|M.ulcerans_Agy99 FQVESRAQMATLPRLKPRVFYDLVVEVALIRPGPIQGGSVHPYIRRRSGI
MAV_4335|M.avium_104 FQVESRAQMATLPRLKPRVFYDLVVEVALIRPGPIQGGSVHPYIRRRNGV
Mflv_4893|M.gilvum_PYR-GCK FQVESRAQMATLPRLKPRMFYDLVVEVALIRPGPIQGGSVHPYIKRRNGQ
Mvan_1530|M.vanbaalenii_PYR-1 FQVESRAQMATLPRLKPRMFYDLVVEVALIRPGPIQGGSVHPYIKRRNGQ
MSMEG_1633|M.smegmatis_MC2_155 FQVESRAQMATLPRLKPREFYDLVVEVALIRPGPIQGGSVHPYIKRRNGQ
MAB_3703c|M.abscessus_ATCC_199 FQVESRAQMATLPRLKPKCFYDLVVEVALIRPGPIQGGSVHPYIRRYNKI
MLBr_01207|M.leprae_Br4923 FQLDGGPMRDLLRRMQPTGFEDIVAVLALYRPGPMGMNAHNDYADRKNNR
TH_0581|M.thermoresistible__bu FQLDGGPMRDLLRRMQPTSFDDIVAVLALYRPGPMGMNAHNDYADRKNGR
**::. . * *::* * *:*. :** ****: .: : * * .
Mb3405c|M.bovis_AF2122/97 DP-VIYEHPSMAP----ALRKTLGVPLFQEQLMQLAVDCAGFSAAEADQL
Rv3370c|M.tuberculosis_H37Rv DP-VIYEHPSMAP----ALRKTLGVPLFQEQLMQLAVDCAGFSAAEADQL
MMAR_1158|M.marinum_M DP-VVYEHPSMES----ALRKTLGVPLFQEQLMQLAVDCAGFSAAEADQL
MUL_0923|M.ulcerans_Agy99 DP-VVYEHPSMES----ALRKTLGVPLFQEQLMQLAVDCAGFSAAEADQL
MAV_4335|M.avium_104 DP-VLYDHPSMEP----ALRKTLGVPLFQEQLMQLAVDCAGFSAAEADQL
Mflv_4893|M.gilvum_PYR-GCK EP-VTYEHPSMER----ALRKTLGVPLFQEQLMQLAVDCAGFSAAEADQL
Mvan_1530|M.vanbaalenii_PYR-1 EA-VTYDHPSMES----ALRKTLGVPLFQEQLMQLAVDCAGFTPAEADQL
MSMEG_1633|M.smegmatis_MC2_155 EP-VTYDHPSMEP----ALKKTLGVPLFQEQLMQLAVDCAGFSAAEADQL
MAB_3703c|M.abscessus_ATCC_199 DKDWQHDHPSMAA----ALDKTLGVPLFQEQLMQLAVDVAGFSPAESDQL
MLBr_01207|M.leprae_Br4923 QV-IKPIHPELEEPLREILAETYGLIVYQEQIMRIAQKVAGYSLARADIL
TH_0581|M.thermoresistible__bu QP-ITPIHPELAEPLEEILAETYGLIVYQEQIMRIAQKVAGYSLARADIL
: **.: * :* *: ::***:*::* . **:: *.:* *
Mb3405c|M.bovis_AF2122/97 RRAMGSKRSTERMRRLRGRFYDGMRALHGAPDEVIDRIYEKLEAFANFGF
Rv3370c|M.tuberculosis_H37Rv RRAMGSKRSTERMRRLRGRFYDGMRALHGAPDEVIDRIYEKLEAFANFGF
MMAR_1158|M.marinum_M RRAIGSKRSTERMRRLRGRFYDGMRALHGAPDEVIDRIYEKLEAFANFGF
MUL_0923|M.ulcerans_Agy99 RRAIGSKRSTERMRRLRGRFYDGMRALHGAPDEVIDRIYEKLEAFANFGF
MAV_4335|M.avium_104 RRAMGSKRSTERMRRLRGRFYDGMRALHGAPDEVIDRTYEKLEAFANFGF
Mflv_4893|M.gilvum_PYR-GCK RRAMGSKRSTEKMRRLRGRFFEGMAELHGISGDVARRIYEKLEAFANFGF
Mvan_1530|M.vanbaalenii_PYR-1 RRAMGSKRSTEKMRRLRGRFFDGMAELHGVTGDVAQRIYEKLEAFANFGF
MSMEG_1633|M.smegmatis_MC2_155 RRAMGSKRSTAKMRRLRSRFYDGMRERHGITGVVADRIYEKLEAFANFGF
MAB_3703c|M.abscessus_ATCC_199 RRAMGSKRSPERMERLRNRFYEGMRNLHGITGELADRIYEKLYAFANFGF
MLBr_01207|M.leprae_Br4923 RKAMGKKKREVLEKEFEG-FSEGMQAN-GFSVHAIKALWDIILPFADYAF
TH_0581|M.thermoresistible__bu RKAMGKKKREVLDKEYEG-FSEGMKAN-GFSASAIKALWDTVLPFADYAF
*:*:*.*: .. .. * :** * . :: : .**::.*
Mb3405c|M.bovis_AF2122/97 PESHALSFASLVFYSAWFKLHHPAAFCAALLRAQPMGFYSPQSLVADARR
Rv3370c|M.tuberculosis_H37Rv PESHALSFASLVFYSAWFKLHHPAAFCAALLRAQPMGFYSPQSLVADARR
MMAR_1158|M.marinum_M PESHALSFASLVFYSSWFKLHHPAAFCAALLRAQPMGFYSPQSLVADARR
MUL_0923|M.ulcerans_Agy99 PESHALSFASLVFYSSWFKLHHPAAFCAALLRAQPMGFYSPQSLVADARR
MAV_4335|M.avium_104 PESHALSFASLVFYSSWFKLHHPAAFCAALLRAQPMGFYSPQSLVADARR
Mflv_4893|M.gilvum_PYR-GCK PESHSLSFASLVFYSSWFKLHHPAAFCAALLRAQPMGFYSPQTLVADARR
Mvan_1530|M.vanbaalenii_PYR-1 PESHSLSFASLVFYSSWFKLHHPAAFCAALLRAQPMGFYSPQSLVADARR
MSMEG_1633|M.smegmatis_MC2_155 PESHSLSFASLVFYSSWFKLHHPAAFCAALLRAQPMGFYSPQSLVADARR
MAB_3703c|M.abscessus_ATCC_199 PESHAQSFASLVFYSSWFKLHHPAAFCAALLRAQPMGFYSPQSLVADARR
MLBr_01207|M.leprae_Br4923 NKSHAAGYGLISYWTAYLKANFAGEYMAGLLTSVGDDKDKAAVYLADCRK
TH_0581|M.thermoresistible__bu NKSHAAGYGLVSYWTAYLKANYPAEYMAGLLTSVRDDKDKAAVYLADCRR
:**: .:. : ::::::* :... : *.** : . .. :**.*:
Mb3405c|M.bovis_AF2122/97 HGVAVHGPCVNASLAHATCENAGTEVRLGLGAVRYLGAELAEKLVAERTA
Rv3370c|M.tuberculosis_H37Rv HGVAVHGPCVNASLAHATCENAGTEVRLGLGAVRYLGAELAEKLVAERTA
MMAR_1158|M.marinum_M HGVLVHGPDVNASLAHATLENAGMQVRLGLGAVRHIGDDLAESLVAERND
MUL_0923|M.ulcerans_Agy99 HGVLVHGPDVNASLAHATLENAGMQVRLGLGAVRHIGDDLAESLVAERND
MAV_4335|M.avium_104 HGVTVHGPDVNASLAHATLENAGTEVRLGLGAVRHIGDDLAEKLVQERKA
Mflv_4893|M.gilvum_PYR-GCK HGVDVHGPDVNASLAHATLENHGLDVRLGLGSIRHIGDELAQRLVEDRKL
Mvan_1530|M.vanbaalenii_PYR-1 HGVVVHGPDVNAGLAHATLENHGLDVRLGLGGVRHIGDELAERLVGERKA
MSMEG_1633|M.smegmatis_MC2_155 HGVTVHGPDVNASLAHAGLENRGLDVRLGLGSVRHIGDDLAQRIVDEREA
MAB_3703c|M.abscessus_ATCC_199 HGVTVHGPDVNASLSYATLENAGLEVRIGLGAVRHIGDDLAQAIVEERKV
MLBr_01207|M.leprae_Br4923 FGITVLPPDVNESVLDFAS--VGADIRYGLGAVRNVGANVVGSLIKTRNA
TH_0581|M.thermoresistible__bu LGITVLPPDVNESAVNFTS--VGKDIRYGLGAVRNVGANVVSSLIATRTE
*: * * ** . * ::* ***.:* :* ::. :: *
Mb3405c|M.bovis_AF2122/97 NGPFTSLPDLTSRVQLSVP---QVEALATAGALGCFGMSRREALWAAGAA
Rv3370c|M.tuberculosis_H37Rv NGPFTSLPDLTSRVQLSVP---QVEALATAGALGCFGMSRREALWAAGAA
MMAR_1158|M.marinum_M NGPFASLLNLTSRVQLSVP---QTEALATAGALGCFGMSRREALWAAGAA
MUL_0923|M.ulcerans_Agy99 NGPFASLLNLTSRVQLSVP---QTEALATTGALGCFGMSRREALWAAGAA
MAV_4335|M.avium_104 NGPFASLLDLTARLQLSVP---QTEALATAGAFGCFGMSRREALWAAGAA
Mflv_4893|M.gilvum_PYR-GCK NGPFVSLTDLTRRVQLTVP---QTEALATAGALGCFGITRREGLWAAGAA
Mvan_1530|M.vanbaalenii_PYR-1 HGPFTSLTDLTRRVQLSVP---QTEALATAGALGCFGITRREGLWAAGAA
MSMEG_1633|M.smegmatis_MC2_155 NGQFTSLLDLTSRVQLTVP---QTEALATAGALGCFGITRREGLWAAGAA
MAB_3703c|M.abscessus_ATCC_199 RGPFVSLLDLTGRIQLTTA---QVEALATGGALGCFDMSRREALWVAGAA
MLBr_01207|M.leprae_Br4923 KGKFADFSDYLNKIDITSCNKKVTESLIKAGAFDSLGHSRK-GLFLVHAD
TH_0581|M.thermoresistible__bu KGKFTDFSDYLNKIDISVCNKKVTESLIKAGAFDSLGHPRK-GLFLVHGD
.* *..: : ::::: .*:* . **:..:. .*: .*: . .
Mb3405c|M.bovis_AF2122/97 ATGRPDRLPGVGSSSHIPALPGMSELELAAADVWATGVSPDSYPTQFLRA
Rv3370c|M.tuberculosis_H37Rv ATGRPDRLPGVGSSSHIPALPGMSELELAAADVWATGVSPDSYPTQFLRA
MMAR_1158|M.marinum_M ATQRPDRLPGVGSSSHIPTLPGMSELELAASDVWATGISPDSYPTQFLRA
MUL_0923|M.ulcerans_Agy99 ATQRPDRLPGVGSSSHIPTLPGMSELELAASDVWATGISPDSYPTQFLRA
MAV_4335|M.avium_104 ATQRPDRLPGVGSSSHIPALPGMSELELAAADVWATGISPDSYPTQFLRD
Mflv_4893|M.gilvum_PYR-GCK ATERPDRLPGVGSSSQVPSLPGMTELELTAADVWATGVSPDRYPTEFLRE
Mvan_1530|M.vanbaalenii_PYR-1 ATERPDRLPGVGSSSHVPSLPGMTELELTVADVWATGVSPDRYPTEFLRE
MSMEG_1633|M.smegmatis_MC2_155 ATQRPDRLPGVGSSTHIPPLPGMSALELSAADVWATGISPDSYPTEYLRK
MAB_3703c|M.abscessus_ATCC_199 AAQRPDRLPGVGVSSRIPALPEMGEVTRAAADVWATGVSPDSYPTQFLRA
MLBr_01207|M.leprae_Br4923 AVDSVLGTKKAEAIGQFDLFGG---TDGGTDAVFTIKVPDDEWEDKHKLA
TH_0581|M.thermoresistible__bu AVESVLGTKKAEAMGQFDLFGGDGSSEDSLDSVFTIKVPDEEWDDKHKLA
*. . :. : *:: :. : : :.
Mb3405c|M.bovis_AF2122/97 DLDAMG----------------------VLPAERLGSVSDGDRVLIAGAV
Rv3370c|M.tuberculosis_H37Rv DLDAMG----------------------VLPAERLGSVSDGDRVLIAGAV
MMAR_1158|M.marinum_M DLDALG----------------------VLAAAALLSVPDGERVLIAGAV
MUL_0923|M.ulcerans_Agy99 DLDALG----------------------VLAAAALLSVPDGERVLIAGAV
MAV_4335|M.avium_104 DLDAMG----------------------VVPAARLGSVPDGDRVLIAGAV
Mflv_4893|M.gilvum_PYR-GCK DLDAMG----------------------VVPADRLLSVRDGTRVLVAGAV
Mvan_1530|M.vanbaalenii_PYR-1 DLDAMG----------------------VVPADQLLSLPDGTRVLVAGAV
MSMEG_1633|M.smegmatis_MC2_155 HLDSLG----------------------VVPTDRLLDVADGTRILVAGAV
MAB_3703c|M.abscessus_ATCC_199 RLDAMG----------------------VVPAKGLFGVPDGSRVLVAGAV
MLBr_01207|M.leprae_Br4923 LEREMLGLYVSGHPLNGVAHLLAAQVDTAIPTILDGGVSNDTQVRVGGIL
TH_0581|M.thermoresistible__bu LEREMLGLYVSGHPLEGVAHLLANQVDTQIPAILDGEVPNDAQVTVGGIL
: :.: : :. :: :.* :
Mb3405c|M.bovis_AF2122/97 THRQRPATAQGVTFIN--LEDETGMVNVLCTPGVWARHRKLAHTAPALLI
Rv3370c|M.tuberculosis_H37Rv THRQRPATAQGVTFIN--LEDETGMVNVLCTPGVWARHRKLAHTAPALLI
MMAR_1158|M.marinum_M THRQRPATAQGVTFIN--LEDETGMVNVLCTPGVWVRHRKLANTAPALLI
MUL_0923|M.ulcerans_Agy99 THRQRPATAQGVTFIN--LEDETGMVNVLCTPGVWMRHRKLANTAPVLLI
MAV_4335|M.avium_104 THRQRPGTAQGVTFLN--LEDETGMVNVLCTPGVWARHRKLANTAPALLV
Mflv_4893|M.gilvum_PYR-GCK THRQRPATAQGVTFLN--LEDETGMVNVLCSQGIWARHRKLAQTASALVV
Mvan_1530|M.vanbaalenii_PYR-1 THRQRPATAQGVTFMN--LEDETGMVNVLCSQGVWARHRKLAQTASALVV
MSMEG_1633|M.smegmatis_MC2_155 THRQRPATAQGVTFIN--LEDEKGMVNVLCAPGVWSRYRKVAQTAPALIV
MAB_3703c|M.abscessus_ATCC_199 THRQRPATAAGVTFIN--LEDETGMVNVVCSVGLWARYRKLAVTARALII
MLBr_01207|M.leprae_Br4923 AAVNRRVNKNGIPWASAQLDDLTGGIEVMFFPHTYSSYGADIIDDAVVLV
TH_0581|M.thermoresistible__bu NSVNRRVNKNGLPWASAQLEDLTGGIEVLFFPQTYSLYGAEIADDAVVLI
:* . *:.: . *:* .* ::*: : : .:::
Mb3405c|M.bovis_AF2122/97 RGQVQNASGAITVVAERMGRLTLAVGARSRDFR-----------------
Rv3370c|M.tuberculosis_H37Rv RGQVQNASGAITVVAERMGRLTLAVGARSRDFR-----------------
MMAR_1158|M.marinum_M RGQVQNASGAVTVVAERMGRISLAVGSRSRDFR-----------------
MUL_0923|M.ulcerans_Agy99 RGQVQNASSAVTVVAERMGRISLAVGSRSRDFR-----------------
MAV_4335|M.avium_104 RGQVQNASGAITVVAERLGRITLAVGSRSRDFR-----------------
Mflv_4893|M.gilvum_PYR-GCK RGIVQNATGAVTVVADRMGPINMKVASKSRDFR-----------------
Mvan_1530|M.vanbaalenii_PYR-1 RGIVQNATGAVTVVADRMGRLSLRAASKSRDFR-----------------
MSMEG_1633|M.smegmatis_MC2_155 RGIVQNATGAVTVVADRMDAVNLRVGSRSRDFR-----------------
MAB_3703c|M.abscessus_ATCC_199 RGQVQNASGAVSVVADQLRPLDLQIRSTSRDFR-----------------
MLBr_01207|M.leprae_Br4923 NAKVVVRDDRIALIANQLVVPDFSNVQEDRPLAVSLLTRQCTFDKVNALK
TH_0581|M.thermoresistible__bu KAKVAARDDRIALIAHELIVPDFSNASADRPIAVSLPTRQCTLDKVTALK
.. * . ::::*..: : .* :
Mb3405c|M.bovis_AF2122/97 --------------------------------------------------
Rv3370c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1158|M.marinum_M --------------------------------------------------
MUL_0923|M.ulcerans_Agy99 --------------------------------------------------
MAV_4335|M.avium_104 --------------------------------------------------
Mflv_4893|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1530|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1633|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3703c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01207|M.leprae_Br4923 QVLARHPGTSQVHLRLISGDRITTLELDQSLRVTSSPALMGDLKALLGPG
TH_0581|M.thermoresistible__bu QVLTNHPGTAQVHLRLINGDRITTLELDPSLRVTPSLELMGDLKALLGPG
Mb3405c|M.bovis_AF2122/97 ----
Rv3370c|M.tuberculosis_H37Rv ----
MMAR_1158|M.marinum_M ----
MUL_0923|M.ulcerans_Agy99 ----
MAV_4335|M.avium_104 ----
Mflv_4893|M.gilvum_PYR-GCK ----
Mvan_1530|M.vanbaalenii_PYR-1 ----
MSMEG_1633|M.smegmatis_MC2_155 ----
MAB_3703c|M.abscessus_ATCC_199 ----
MLBr_01207|M.leprae_Br4923 CLGD
TH_0581|M.thermoresistible__bu CLG-