For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GIASSTVDPALRTRAFARVIGPFVATVTTIIAVRAGDLGFLGDSFFRDPMWSWLFGALLFFCGLMIIAFH QFWRGAAAVIISLFGWFLALRGLVLLAFPGLMRAGVDASVPAVGPVRGFFLFMAVVGVYLTYVGWIRKR*
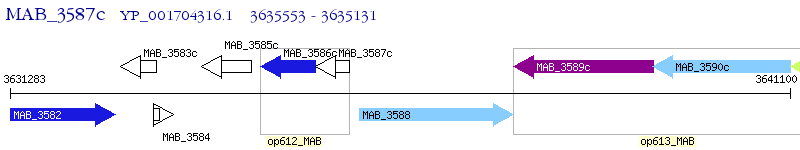
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3587c | - | - | 100% (140) | hypothetical protein MAB_3587c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_5360 | - | 8e-27 | 39.39% (132) | hypothetical protein MMAR_5360 |
| M. avium 104 | MAV_2504 | - | 2e-20 | 40.00% (120) | hypothetical protein MAV_2504 |
| M. smegmatis MC2 155 | MSMEG_3937 | - | 7e-23 | 36.29% (124) | hypothetical protein MSMEG_3937 |
| M. thermoresistible (build 8) | TH_4565 | - | 3e-21 | 39.39% (132) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_4979 | - | 4e-27 | 39.39% (132) | hypothetical protein MUL_4979 |
| M. vanbaalenii PYR-1 | Mvan_4730 | - | 4e-41 | 56.64% (143) | hypothetical protein Mvan_4730 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5360|M.marinum_M ---MSVSLDSQARTRMFCRVLGPLLVIVDVTAVARASDMQNLLSQFEANS
MUL_4979|M.ulcerans_Agy99 ---MSVSLDSQARTRMFCRVLGPLLVIVDVTAVARASDMQNLLSQFEANS
MAV_2504|M.avium_104 ---------------MFARVLGPFLVVFTATTVARASDMRKLLSDFDSNS
MSMEG_3937|M.smegmatis_MC2_155 ---MSTAFHVENRTQGFARVLGPYLVVVAVVALARASEIPGLVTDFGKNP
TH_4565|M.thermoresistible__bu ---MEISQDSALATRRFARVLGPYMVIVPLAAAARPSDTQAVVGAIDASG
MAB_3587c|M.abscessus_ATCC_199 MGIASSTVDPALRTRAFARVIGPFVATVTTIIAVRAGDLGFLGDSFFRDP
Mvan_4730|M.vanbaalenii_PYR-1 MSAPVTTTDRSRRTRSFARVLGPFIAVVPAIVAIRAGSIGSQLSSFSADP
*.**:** :. . *... : .
MMAR_5360|M.marinum_M LWTWVTGAFVLVLGLVMVAGHQYWHGAAAIIVSVLGWIVTLRGVLLLAFP
MUL_4979|M.ulcerans_Agy99 LWTWVTGAFVLVLGLVMVASHQYWHGAAAIIVSVLGWIVTLRGVLLLAFP
MAV_2504|M.avium_104 ALPLVTGGFLLLASLVILGLHQCWRGAPAVTVSVVGWVFAVRALLLMAFP
MSMEG_3937|M.smegmatis_MC2_155 IMSWVSGAFVLLSGLTVVALHQRWHGVAAIVVSVLGWLATLKGVLLLAVP
TH_4565|M.thermoresistible__bu VWQLLTGVFALLAGLVIVGLHEHRRGPAAIIVSVVGWLAVLEGLVLLVVP
MAB_3587c|M.abscessus_ATCC_199 MWSWLFGALLFFCGLMIIAFHQFWRGAAAVIISLFGWFLALRGLVLLAFP
Mvan_4730|M.vanbaalenii_PYR-1 MWSWSLGALLFLCGVFIIAFHQYWRSVAAVIISLFGWFLLLRGLALLTVP
* : :. .: ::. *: :. .*: :*:.**. :..: *:..*
MMAR_5360|M.marinum_M KAFVSAANSMIG----AQAVWVSLCIVFALLGLYLTYVGWAPAASGPSQH
MUL_4979|M.ulcerans_Agy99 KAFVSAANSMIG----AQSVWVSLCIVFALLGLYLTYVGWAPAASGPSQH
MAV_2504|M.avium_104 HAFMAAADAAVR----MTALWVSGDVVVGVVGLYLAYVGWKPVTS-PSTP
MSMEG_3937|M.smegmatis_MC2_155 QSYAAFGESMVR----AAMWWQVLMVAVGLLGLFLTYAGWAPANE-PRTA
TH_4565|M.thermoresistible__bu RAYLALADWLVG----STVLLWVVMLVAAAIGAYLTYVGWIGERSVAQEP
MAB_3587c|M.abscessus_ATCC_199 GLMRAGVDASVP----AVGPVRGFFLFMAVVGVYLTYVGWIRKR------
Mvan_4730|M.vanbaalenii_PYR-1 QLIVEGAESATQTSDTAVDVVRVGFGLLAVCGLYLAYVGWVKKPD-----
: . * :*:*.**
MMAR_5360|M.marinum_M EAAA--SRDLPRAA
MUL_4979|M.ulcerans_Agy99 EAAA--SRDLPRAA
MAV_2504|M.avium_104 ATTS--TPDVPRAA
MSMEG_3937|M.smegmatis_MC2_155 RTFGGPAPDLPSAA
TH_4565|M.thermoresistible__bu GSRA-------RA-
MAB_3587c|M.abscessus_ATCC_199 --------------
Mvan_4730|M.vanbaalenii_PYR-1 --------------