For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSPREYDESDARIRPGRRSRPRTKIRPDHADAAPAMVVTVDRGRWGCVLHDDPAQQVTAMRARELGRTPI VVGDTVSLVGDLSGRPDTLARIVRLEERRTVLRRTADDTDPFERIFVANADQLLIVVAVADPPPRTGLVE RALIAAYAGGLTPILALTKTDLADPDSFAAQFADLNIRVIGAGRDEPLAPIIEVLTGRLTALLGHSGVGK STLVNRLVPEAGRATGEVTGVGKGRHTSTQSVALPLYPSGWVIDTPGIRSFGLAHIRPDDVLAAFSDLSE AIEDCPRGCQHLGPPADPECALDELEGASAKRVTAVRQLLSVLTTA
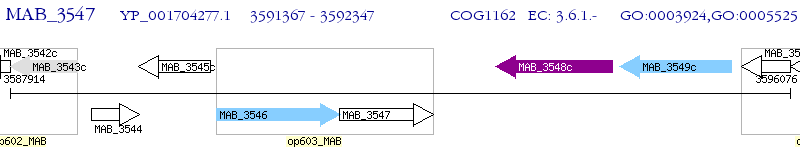
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3547 | - | - | 100% (326) | hypothetical protein MAB_3547 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3257 | - | 1e-137 | 73.48% (328) | hypothetical protein Mb3257 |
| M. gilvum PYR-GCK | Mflv_4696 | - | 1e-139 | 76.16% (323) | GTPase EngC |
| M. tuberculosis H37Rv | Rv3228 | - | 1e-137 | 73.48% (328) | hypothetical protein Rv3228 |
| M. leprae Br4923 | MLBr_00791 | - | 1e-136 | 74.21% (318) | hypothetical protein MLBr_00791 |
| M. marinum M | MMAR_1319 | - | 1e-139 | 74.31% (327) | hypothetical protein MMAR_1319 |
| M. avium 104 | MAV_4184 | - | 1e-141 | 76.00% (325) | GTPase EngC |
| M. smegmatis MC2 155 | MSMEG_1889 | - | 1e-143 | 77.67% (318) | putative ribosome small subunit-dependent GTPase RsgA |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2561 | - | 1e-137 | 73.39% (327) | hypothetical protein MUL_2561 |
| M. vanbaalenii PYR-1 | Mvan_1770 | - | 1e-145 | 79.57% (323) | GTPase EngC |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3257|M.bovis_AF2122/97 MRPGD---YDESDVKVRSGRSSRPRTKTRPEHADAEAAMVVSVDRGRWGC
Rv3228|M.tuberculosis_H37Rv MRPGD---YDESDVKVRSGRSSRPRTKTRPEHADAEAAMVVSVDRGRWGC
MLBr_00791|M.leprae_Br4923 MKPDNY--YDESDVKIRSGRGSRPRTKTRPEHADAKAAMVVSVDRGRWGC
MMAR_1319|M.marinum_M MRPGD---YDESDVKIRSGRGSRPRTKTRPEHADAEAAMVVAVDRGRWGC
MUL_2561|M.ulcerans_Agy99 MRPGD---YDESDVKIRSGRGSRPRTKTRPEHADAEAAMVVAVDRGRWGC
MAV_4184|M.avium_104 MRPGD---YDESDVKIRSGRGSRPRTKTRPEHADARAAMVVSVDRGRWGC
Mflv_4696|M.gilvum_PYR-GCK MSPRE---YDETDVRVRPGRGSRPRTKTRPVHADAEHAMVVTVDRGRWGC
Mvan_1770|M.vanbaalenii_PYR-1 MSPRE---YDESDVRIRPGRGSRPRTKTRPEHADAQQAMVVTVDRGRWGC
MSMEG_1889|M.smegmatis_MC2_155 MSARRSLNYDESDVRIRPGRGSRPRTKNRPEHADAQSAMVVTVDRGRWGC
MAB_3547|M.abscessus_ATCC_1997 MSPRE---YDESDARIRPGRRSRPRTKIRPDHADAAPAMVVTVDRGRWGC
* . ***:*.::*.** ****** ** **** ****:********
Mb3257|M.bovis_AF2122/97 VLGGRPDRRITAMRARELGRTPIVVGDDVDVVGDLSGRPDTLARIVRRAP
Rv3228|M.tuberculosis_H37Rv VLGGRPDRRITAMRARELGRTPIVVGDDVDVVGDLSGRPDTLARIVRRAP
MLBr_00791|M.leprae_Br4923 ALDNCPDRRITAMRARELGHTPIVVGDNVEVVGDLSGRPDTLARIVRRLQ
MMAR_1319|M.marinum_M VLGGDPLRRVTAMRARELGRTPIVVGDDVDLVGDLSGRPDTLARIVRRGP
MUL_2561|M.ulcerans_Agy99 VLGGDPLRRVTAMRTRELGRTPIVVGDDVDLVGDLSGRPDTLARIVRRGP
MAV_4184|M.avium_104 VLGGDPDRRVTAMRARELGRTPIVVGDDVDVVGDLSGRPDTLARIVRRGE
Mflv_4696|M.gilvum_PYR-GCK VLGNDPGRRVTAMRARELGRTPIVVGDDVDIVGDLSGRTDTLARIVRRGE
Mvan_1770|M.vanbaalenii_PYR-1 VLGRDPDRRVTAMRARELGRTPIVVGDDVDVVGDLSGRTDTLARIVRRGE
MSMEG_1889|M.smegmatis_MC2_155 ALDRDPDRVVTAMRARELGRTPIVVGDDVDVVGDLSGRPDTLARIVRRGE
MAB_3547|M.abscessus_ATCC_1997 VLHDDPAQQVTAMRARELGRTPIVVGDTVSLVGDLSGRPDTLARIVRLEE
.* * : :****:****:******* *.:*******.********
Mb3257|M.bovis_AF2122/97 RRTVLRRTADDTDPTERVVVANADQLLIVVALADPPPRTGLVDRALIAAY
Rv3228|M.tuberculosis_H37Rv RRTVLRRTADDTDPTERVVVANADQLLIVVALADPPPRTGLVDRALIAAY
MLBr_00791|M.leprae_Br4923 RRTVLRRTADDTDTSERVVVANADQLLIVVALADPPPRTGLVDRALIAAY
MMAR_1319|M.marinum_M RRTVLRRTADDTDPTERVVVANADQLLMVVALADPPPRSGLVDRALIAAY
MUL_2561|M.ulcerans_Agy99 RRTVLRRTADDTDPTERVVVANADQLLMVVALADPPPRSGLVDRALIAAY
MAV_4184|M.avium_104 RRTVLRRTADDTDPTERVVVANADQLLIVVALADPPPRTGLVDRALIAAY
Mflv_4696|M.gilvum_PYR-GCK RRTVLRRTADDTDPTERVVVANADQLLIVVALADPPPRTGLVERALIAAY
Mvan_1770|M.vanbaalenii_PYR-1 RRTVLRRTADDTDPAERVVVANADQLLIVVALADPPPRTGLVERALIAAY
MSMEG_1889|M.smegmatis_MC2_155 RRTVLRRTADDTDPTERVVVANADQLLIVVALADPPPRTGFVERALIAAY
MAB_3547|M.abscessus_ATCC_1997 RRTVLRRTADDTDPFERIFVANADQLLIVVAVADPPPRTGLVERALIAAY
*************. **:.********:***:******:*:*:*******
Mb3257|M.bovis_AF2122/97 AGGLTPILCLTKTDLAPAEPFGKQFADLELTVTAAGVDDPLLAVADLLAG
Rv3228|M.tuberculosis_H37Rv AGGLTPILCLTKTDLAPAEPFGKQFADLELTVTAAGVDDPLLAVADLLAG
MLBr_00791|M.leprae_Br4923 AGGLAPILCLTKTDLAPLEPFAKQFVDLDLTVVTAGVGDPLLAVADLLAG
MMAR_1319|M.marinum_M TGGLTPILCLTKTDLAPPEPFAEQFLELDLQVVAAGRDDPLLAVADLLDG
MUL_2561|M.ulcerans_Agy99 TGGLTPILCLTKTDLAPPEPFAEQFLELDLQVVAAGRDDPLLAVADLLDG
MAV_4184|M.avium_104 AGGLVPILCLTKTDLAPPEPFAQQFADLDLTVVAAGRDDPLPAVADLLAG
Mflv_4696|M.gilvum_PYR-GCK AGGLVPILCLTKTDLAPAEPFAEQFRDLDLTVLTAGRDDALETVDALLAN
Mvan_1770|M.vanbaalenii_PYR-1 AGGLVPILCLTKTDLAPAEPFAAQFRDLDLVIVTAGRDDPLEAVGSLLAD
MSMEG_1889|M.smegmatis_MC2_155 AGGLKPVLCLTKSDLAAPEAFAAQFADLDLTIVTAGRDDPIEPVRPLLTG
MAB_3547|M.abscessus_ATCC_1997 AGGLTPILALTKTDLADPDSFAAQFADLNIRVIGAGRDEPLAPIIEVLTG
:*** *:*.***:*** :.*. ** :*:: : ** .:.: .: :* .
Mb3257|M.bovis_AF2122/97 KITVLLGHSGVGKSTLVNRLVPEADRAVGEVTEIGRGRHTSTRSVALPLG
Rv3228|M.tuberculosis_H37Rv KITVLLGHSGVGKSTLVNRLVPEADRAVGEVTEIGRGRHTSTRSVALPLG
MLBr_00791|M.leprae_Br4923 KITVLLGHSGVGKSTLVNRLVPEANRSVGEVTDTGRGRHTSTQSVALPL-
MMAR_1319|M.marinum_M KTTVLLGHSGVGKSTLVNRLVPEADRAVGEVTEIGRGRHTSTQSVALPLE
MUL_2561|M.ulcerans_Agy99 KTTVLLGHSGVGKSTLVNRLVPEADRAVGEVTEIGRGRHTSTQSVALPLE
MAV_4184|M.avium_104 KITVLLGHSGVGKSTLVNRLVPDADRAVGAVTEIGKGRHTSTQSVALPLT
Mflv_4696|M.gilvum_PYR-GCK KVTALLGHSGVGKSTLVNRLVPEADRATGAVTDIGRGRHTSTQSVALPLL
Mvan_1770|M.vanbaalenii_PYR-1 KVTALLGHSGVGKSTLVNRLVPEAERATGEVTDVGKGRHTSTQSVALPLR
MSMEG_1889|M.smegmatis_MC2_155 QVTVFLGHSGVGKSTLVNRLIPEADRATGEVSGVGKGKHTSTQSVALPLD
MAB_3547|M.abscessus_ATCC_1997 RLTALLGHSGVGKSTLVNRLVPEAGRATGEVTGVGKGRHTSTQSVALPLY
: *.:***************:*:* *:.* *: *:*:****:******
Mb3257|M.bovis_AF2122/97 DTLSGSGWVIDTPGIRSFGLAHIQPDNVLLAFSDLAEATRECPRGCGHMG
Rv3228|M.tuberculosis_H37Rv DTLSGSGWVIDTPGIRSFGLAHIQPDNVLLAFSDLAEATRECPRGCGHMG
MLBr_00791|M.leprae_Br4923 ---EGSGWVIDTPGIRSFGLAHIRPDDVVLAFSDLTEATEDCPRGCRHIG
MMAR_1319|M.marinum_M TDGQGGGWVIDTPGIRSFGLAHIQPDDVLLAFTDLAEAIGDCPRGCGHMG
MUL_2561|M.ulcerans_Agy99 TDGQGGGWVIDTPGIRSLGLAHIQPDDVLLAFTDLAEAIGDCPRGCGHMG
MAV_4184|M.avium_104 ERLGTTGWVIDTPGIRSFGLAHIQPDDVVMAFSDLAEAIADCPRGCGHMG
Mflv_4696|M.gilvum_PYR-GCK ----HGGWVVDTPGIRSFGLAHIAPDDVVMAFSDLADAIDGCPRGCGHMG
Mvan_1770|M.vanbaalenii_PYR-1 ----LGGWVIDTPGIRSFGLAHIEPDDVVLAFSDLAEAIDDCPRGCGHMG
MSMEG_1889|M.smegmatis_MC2_155 ----GDGWVIDTPGIRSFGLAHIEPDDVILAFSDLADTIEDCPRGCGHMG
MAB_3547|M.abscessus_ATCC_1997 ----PSGWVIDTPGIRSFGLAHIRPDDVLAAFSDLSEAIEDCPRGCQHLG
***:*******:***** **:*: **:**::: ***** *:*
Mb3257|M.bovis_AF2122/97 PPADPECALDTLSGPAARRAAAARRLLAVLSQT------
Rv3228|M.tuberculosis_H37Rv PPADPECALDTLSGPAARRAAAARRLLAVLSQT------
MLBr_00791|M.leprae_Br4923 PPADPECTLDNISGPAVRRVAATRRLLAVLRET------
MMAR_1319|M.marinum_M PPADPECVLDSLSGTAVRRVAAARRLLTALRET------
MUL_2561|M.ulcerans_Agy99 PPADPECVLDSLSGTAVRRLAAARRLLTALRET------
MAV_4184|M.avium_104 PPADPECALDTLTGPAARRVAAARRLLDALNQTS-----
Mflv_4696|M.gilvum_PYR-GCK PPADPGCALDALTGAAEGRVHAARRLLAALRES------
Mvan_1770|M.vanbaalenii_PYR-1 PPADPECALDELTGAAQGRVQAARRLLAALREG------
MSMEG_1889|M.smegmatis_MC2_155 PPADPECALDALTGPAADRAAAARRLLAVLKDTRRAMNI
MAB_3547|M.abscessus_ATCC_1997 PPADPECALDELEGASAKRVTAVRQLLSVLTTA------
***** *.** : *.: * *.*:** .*