For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AEEVHAEIVASVLEVLVKKGDAIAEGDTIVLLESMKMEIPVLAEVAGTIGDVSVSVGDVIQAGDLIAVID D*
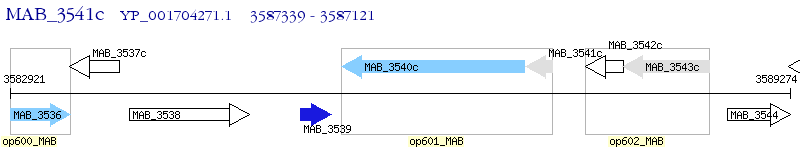
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3541c | - | - | 100% (72) | putative acetyl-CoA carboxylase biotin carboxyl carrier protein subunit |
| M. abscessus ATCC 19977 | MAB_3643 | - | 2e-09 | 42.65% (68) | bifunctional protein acetyl-/propionyl-CoA carboxylase (alpha |
| M. abscessus ATCC 19977 | MAB_2066c | - | 1e-08 | 35.38% (65) | acetyl/propionyl carboxylase alpha subunit AccA2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3247c | TB7.3 | 2e-24 | 77.14% (70) | putative acetyl-CoA carboxylase biotin carboxyl carrier |
| M. gilvum PYR-GCK | Mflv_4687 | - | 4e-26 | 82.86% (70) | putative acetyl-CoA carboxylase biotin carboxyl carrier |
| M. tuberculosis H37Rv | Rv3221c | TB7.3 | 2e-24 | 77.14% (70) | putative acetyl-CoA carboxylase biotin carboxyl carrier |
| M. leprae Br4923 | MLBr_00802 | - | 3e-24 | 77.14% (70) | putative acetyl-CoA carboxylase biotin carboxyl carrier |
| M. marinum M | MMAR_1336 | - | 7e-24 | 74.29% (70) | hypothetical protein MMAR_1336 |
| M. avium 104 | MAV_4169 | - | 2e-25 | 80.00% (70) | putative acetyl-CoA carboxylase biotin carboxyl carrier |
| M. smegmatis MC2 155 | MSMEG_1917 | - | 3e-24 | 77.46% (71) | putative acetyl-CoA carboxylase biotin carboxyl carrier |
| M. thermoresistible (build 8) | TH_0667 | - | 2e-26 | 80.28% (71) | BIOTINYLATED PROTEIN TB7.3 |
| M. ulcerans Agy99 | MUL_2543 | - | 5e-24 | 74.29% (70) | hypothetical protein MUL_2543 |
| M. vanbaalenii PYR-1 | Mvan_1780 | - | 2e-26 | 82.86% (70) | putative acetyl-CoA carboxylase biotin carboxyl carrier |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1336|M.marinum_M --------------------------------------------------
MUL_2543|M.ulcerans_Agy99 MPWLALLCLRSRFLRSRLAMMNLRSWSMQGGQVPPGLSQVSHAGVSATAL
Mb3247c|M.bovis_AF2122/97 --------------------------------------------------
Rv3221c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00802|M.leprae_Br4923 --------------------------------------------------
MAV_4169|M.avium_104 --------------------------------------------------
Mflv_4687|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1917|M.smegmatis_MC2_155 --------------------------------------------------
TH_0667|M.thermoresistible__bu --------------------------------------------------
Mvan_1780|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3541c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1336|M.marinum_M ---------------------------------MSEDVRAEIVASVLEVV
MUL_2543|M.ulcerans_Agy99 ARAPRPRVALWFDIHEPIDQQPACAMAEPSGVKMSEDVRAEIVASVLEVV
Mb3247c|M.bovis_AF2122/97 ---------------------------------MAEDVRAEIVASVLEVV
Rv3221c|M.tuberculosis_H37Rv ---------------------------------MAEDVRAEIVASVLEVV
MLBr_00802|M.leprae_Br4923 -------------------------MAEMSGVKMAEDVRAEIVASVLEVV
MAV_4169|M.avium_104 -------------------------------MKMAEDVRAEIVASVLEVV
Mflv_4687|M.gilvum_PYR-GCK ---------------------------------MAEDVRAEIVASVLEVV
MSMEG_1917|M.smegmatis_MC2_155 -------------------------------MKMAEDVRAEIVASVLEVV
TH_0667|M.thermoresistible__bu ---------------------------------MAEDVRAEIVASVLEVV
Mvan_1780|M.vanbaalenii_PYR-1 ---------------------------------MAEDVRAEIVASVLEVV
MAB_3541c|M.abscessus_ATCC_199 ---------------------------------MAEEVHAEIVASVLEVL
*:*:*:**********:
MMAR_1336|M.marinum_M VSEGDQIGKGDIVVLLESMKMEIPVLAEIGGTVSKVSVAVGDVIQAGDLI
MUL_2543|M.ulcerans_Agy99 VSEGDQIGKGDIVVLLESMKMEIPVLAEIGGTVSKVSVAVGDVIQAGDLI
Mb3247c|M.bovis_AF2122/97 VNEGDQIDKGDVVVLLESMKMEIPVLAEAAGTVSKVAVSVGDVIQAGDLI
Rv3221c|M.tuberculosis_H37Rv VNEGDQIDKGDVVVLLESMKMEIPVLAEAAGTVSKVAVSVGDVIQAGDLI
MLBr_00802|M.leprae_Br4923 VSEGDQIGKGDVLVLLESMKMEIPVLAGVAGIVSKVSVSVGDVIQAGDLI
MAV_4169|M.avium_104 VNEGDQIGKGDTLVLLESMKMEIPVLAEVGGTVSKVSVSVGDVIQAGDLI
Mflv_4687|M.gilvum_PYR-GCK VHEGDQIGEGDTLVLLESMKMEIPVLAEVAGTVSKVSVSVGDVIQAGDLI
MSMEG_1917|M.smegmatis_MC2_155 VHEGDQIGEGDTLVLLESMKMEIPVLAEVAGTVTKVNVAEGDVIQAGHLI
TH_0667|M.thermoresistible__bu VQEGDQIGEGDTLVLLESMKMEIPVLAEVAGTVSKVNVAVGDVIQAGDLI
Mvan_1780|M.vanbaalenii_PYR-1 VREGDQIGEGDTLVLLESMKMEIPVLAEVAGTISKVSVSVGDVIQAGDLI
MAB_3541c|M.abscessus_ATCC_199 VKKGDAIAEGDTIVLLESMKMEIPVLAEVAGTIGDVSVSVGDVIQAGDLI
* :** * :** :************** .* : .* *: *******.**
MMAR_1336|M.marinum_M AVIS-
MUL_2543|M.ulcerans_Agy99 AVIS-
Mb3247c|M.bovis_AF2122/97 AVIS-
Rv3221c|M.tuberculosis_H37Rv AVIS-
MLBr_00802|M.leprae_Br4923 AVIS-
MAV_4169|M.avium_104 AVIS-
Mflv_4687|M.gilvum_PYR-GCK AVIS-
MSMEG_1917|M.smegmatis_MC2_155 AVID-
TH_0667|M.thermoresistible__bu AVID-
Mvan_1780|M.vanbaalenii_PYR-1 AIIS-
MAB_3541c|M.abscessus_ATCC_199 AVIDD
*:*.